Abstract
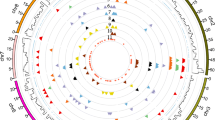
We developed 708 gene-based markers for citrus genome analysis. Sequence-tagged site (STS) primers were designed that were located in conserved exon regions and whose PCR products spanned genomic introns. Of these, 79.7 % comprised cleaved amplified polymorphic sequence markers. The gene-based markers and their annotation and position on Clementine scaffolds ver. 1.0 permitted comparison of the genetic map and the Clementine genome sequence. The 708 gene-based markers were used to construct a genetic map using the 87 progenies (AG population) from the cross between ‘Okitsu 46 gou’ (‘Sweet Spring’ (‘Ueda unshiu’ (Citrus unshiu) × Hassaku (Citrus hassaku Hort. ex Tanaka)) × ‘Trovita’ orange) × ‘Kankitsu Chukanbohon Nou 5 gou’ (‘Lee’ (Citrus clementina × tangelo) × Citrus kinokuni). The markers were integrated using common STSs on different phase maps in cross-pollination mode. The integrated map (AGI map) comprised 706 loci, including two morphological traits, and spanned 990.9 centimorgans (cM) with an average marker distance of 1.40 cM. These markers formed nine linkage groups (LGs) (corresponding to citrus physical chromosomes): LG-01 to LG-09 corresponded to Scaffold_01, Scaffold_07, Scaffold_09, Scaffold_06, Scaffold_03, Scaffold_02, Scaffold_04, Scaffold_08, and Scaffold_05, respectively. LG-08 and LG-09 contained morphological traits controlling embryo color and seedlessness. Eighty-eight loci comprised three or more alleles on the AGI map; 36.4 % of them were related to transcription factors and DNA-binding proteins. The 708 gene-based markers and the AGI map are valuable for integrating various citrus genetic maps, alignment of genomic sequences, chromosome assignment, and understanding the diversity of citrus germplasms.




Similar content being viewed by others
References
Aubourg S, Lecharny A, Bohlmann J (2002) Genomic analysis of the terpenoid synthase (AtTPS) gene family of Arabidopsis thaliana. Mol Genet Genomics 267:730–745
Blanc G, Barakat A, Guyot R, Cooke R, Delseny M (2000) Extensive duplication and reshuffling in the Arabidopsis genome. Plant Cell 12:1093–1101
Cai Q, Guy CG, Moore GA (1994) Extension of the linkage map in Citrus using random amplified polymorphic DNA (RAPD) markers and RFLP mapping of cold-acclimation-responsive loci. Theor Appl Genet 89:606–614
Cristofani M, Machado MA, Grattapaglia D (1999) Genetic linkage maps of Citrus sunki Hort. Ex. Tan. and Poncirus trifoliata (L.) Raf. and mapping of citrus tristeza virus resistance gene. Euphytica 109:25–32
De Simone M, Russo MP, Puleo G, Marsan PA, Lorenzoni C, Marocco A, Recupero GR (1998) Construction of genetic maps for Citrus aurantium and C. latipes based on AFLP, RAPD and RFLP markers. Fruits 53:383–390
Dellaporta SL, Wood J, Hicks JB (1983) A plant DNA minipreparation: version II. Plant Mol Biol Rep 1:19–21
Durham RE, Liou PC, Gmitter FG Jr, Moore GA (1992) Linkage of restriction fragment length polymorphisms and isozymes in Citrus. Theor Appl Genet 84:39–48
Endo T, Shimada T, Fujii H, Kobayashi Y, Araki T, Omura M (2005) Ectopic expression of an FT homolog from Citrus confers an early flowering phenotype on trifoliate orange (Poncirus trifoliata L. Raf.). Transgenic Res 14:703–712
Endo T, Shimada T, Fujii H, Omura M (2006) Cloning and characterization of 5 MADS-box cDNAs isolated from citrus fruit tissue. Sci Hort 109:315–321
Fang DQ, Federici CT, Roose ML (1997) Development of molecular markers linked to a gene controlling fruit acidity in citrus. Genome 40:841–849
Fang DQ, Federici CT, Roose ML (1998) A high-resolution map of the citrus tristeza virus resistance gene region in Poncirus trifoliate (L.). Raf Genetic 150:883–890
Forment J, Gadea L, Huerta L et al (2005) Development of a citrus genome-wide EST collection and cDNA microarray as resources for genomic studies. Plant Mol Biol 57:375–391
Fujii H, Kita M, Shimada T, Endo T, Omura M (2003) Expressed sequence tags from citrus albedo at the initiation stage of rind peeling. Bull Natl Inst Fruit Tree Sci 2:127–143
Fujii H, Ogata T, Sonoda K, Sugiyama A, Shimada T, Endo T, Shimizu T, Omura M (2007a) Development of software to provide automatically structural and functional annotation for EST utilizing genome information of Arabidopsis thaliana. Ja Hort Res Suppl 6:36
Fujii H, Ogata T, Sonoda K, Sugiyama A, Shimada T, Endo T, Shimizu T, Omura M (2007b) Profiling ethylene-responsive genes in mature mandarin fruit using a citrus 22 K oligoarray. Plant Sci 173:340–348
Fujii H, Shimada T, Sugiyama A, Nishikawa F, Endo T, Nishikawa F, Nakano M, Ikoma Y, Shimizu T, Omura M (2008) Profiling gibberellin (GA3)-responsive genes in mature mandarin fruit using a citrus 22 K oligoarray. Sci Hort 116:291–298
Fujii H, Shimada T, Nonaka K, Kita M, Kuniga T, Endo T, Ikoma Y, Omura M (2013) High-throughput genotyping in citrus accessions using an SNP genotyping array. Tree Genetics & Genomes 9:145–153
Garcia R, Asins MJ, Forner J, Carbonell EA (1999) Genetic analysis of apomixes in Citrus and Poncirus by molecular markers. Theor Appl Genet 99:511–518
Gmitter FG Jr, Xiao SY, Huang S, Hu XL, Garnsey SM, Deng Z (1996) A localized linkage map of the citrus tristeza virus resistance gene region. Theor Appl Genet 92:688–695
Goff SA, Ricke D, Lan TH et al (2002) A draft sequence of the rice genome (Oryza sativa L. ssp. japonica). Science 296:92–100
Hisada S, Moriguchi T, Hidaka T, Koltunow AM, Akihama T, Omura M (1996) Random sequencing of sweet orange (Citrus sinensis Osbeck) cDNA library derived from young seeds. J Japan Soc Hort Sci 65:487–495
Hisada T, Akihama T, Endo T, Moriguchi T, Omura M (1997) Expressed sequence tags of Citrus fruit during rapid cell development phase. J Amer Soc Hort Sci 122:808–812
Hisada S, Kita M, Endo-Inagaki T, Omura M, Moiguchi T (1999) Refinement of cDNA clone expression analysis in random sequencing from the rapid cell development phase of citrus fruit. J Plant Physiol 155:699–705
Jarrell DC, Roose ML, Traugh SN, Kupper RS (1992) A genetic map of citrus based on the segregation of isozymes and RFLPs in an intergeneric cross. Theor Appl Genet 84:49–56
Kato M, Ikoma Y, Matsumoto H, Sugiura M, Hyodo H, Yano M (2004) Accumulation of carotenoids and expression of carotenoid biosynthetic genes during maturation in citrus fruit. Plant Physiol 134:824–837
Kijas JM, Thomas MR, Fowler JSC, Roose ML (1997) Integration of trinucleotide microsatellites into a linkage map of Citrus. Theor Appl Genet 94:701–706
Kita M, Endo-Inagaki T, Moriguchi T, Omura M (2000a) cDNA catalogs expressed in albedo of citrus fruit: a comparative analysis of cDNA libraries from pulp and albedo of Satsuma mandarin (Citrus unshiu Marc.). Acta Hort 521:179–183
Kita M, Hisada S, Endo-Inagaki T, Omura M, Moriguchi T (2000b) Changes in the levels of mRNAs for the putative cell growth-related genes in rind (albedo and flavedo) during citrus fruit development. Plant Cell Physiol 19:582–587
Kita M, Hirata Y, Moriguchi T, Endo-Inagaki T, Matsumoto R, Hasegawa S, Suhayda CG, Omura M (2000c) Molecular cloning and characterization of a novel gene encoding limonoid UDP-glucosyltransferase in Citrus. FEBS Lett 469:173–178
Komatsu A, Takanokura Y, Omura M, Akihama T (1996) Cloning and molecular analysis of cDNAs encoding three sucrose phosphate synthase isoforms from a citrus fruit (Citrus unshiu Marc.). Mol Gen Genet 252:346–351
Komatsu A, Moriguchi T, Koyama K, Omura M, Akihama T (2002) Analysis of sucrose synthase genes in citrus suggests different roles and phylogenetic relationships. J Exp Bot 53:61–71
Konieczny K, Ausubel FM (1993) A procedure for mapping Arabidopsis mutations using co-dominant ecotype-specific PCR-based markers. Plant J 4:403–410
Ling P, Duncan LW, Deng Z, Dunn D, Hu X, Huang S, Gmitter FG Jr (2000) Inheritance of citrus nematode resistance and its linkage with molecular markers. Theor Appl Genet 100:1010–1017
Liou P-C, Gmitter FG Jr, Moore GA (1996) Characterization of the Citrus genome through analysis of restriction fragment length polymorphisms. Theor Appl Genet 92:425–435
Luro FL, Costantino G, Terol J, Argout X, Allario T, Wincker P, Talon T, Ollitrault P, Morillon R (2008) Transferability of the EST-SSRs developed on Nules Clementine (Citrus clementina Hort ex Tan) to other Citrus species and their effectiveness for genetic mapping. BMC Genomics 9:287. doi:10.1186/1471-2164-9-287
Martin DM, Aubourg S, Schouwey BM, Daviet L, Schalk M, Toub O, Lund TS, Bohlmann J (2010) Functional annotation, genome organization and phylogeny of the grapevine (Vitis vinifera) terpene synthase gene family based on genome assembly, FLcDNA cloning, and enzyme assays. BMC Plant Biol 10:226. doi:10.1186/1471-2229-10-226
Miranda M, Ikeda F, Endo T, Moriguchi T, Omura M (1997) Comparative analysis on the distribution of heterochromatin in Citrus, Poncirus and Fortunella chromosomes. Chromosome Res 5:86–92
Moretzsohn MC, Leoi L, Proite K et al (2005) A microsatellite-based, gene-rich linkage map for the AA genome of Arachis (Fabaceae). Theor Appl Genet 111:1060–1071
Moriguchi T, Kita M, Hisada S, Endo-Inagaki T, Omura M (1998) Characterization of gene repertories at mature stage of citrus fruit through random sequencing and analysis of redundant metallothionein-like genes expressed during fruit development. Gene 211:221–227
Nakano M, Shimizu T, Fujii H, Shimada T, Endo T, Nesumi H, Kuniga T, Omura M (2008) Marker enrichment and construction of haplotype-specific BAC contigs for the polyembryony genomic region in Citrus. Breed Sci 58:375–383
Ohta S, Endo T, Shimada T, Fujii H, Shimizu T, Kuniga T, Yoshioka T, Nesumi H, Yoshida T, Omura M (2011) PCR primers for marker assisted backcrossing to introduce a CTV resistance gene from Poncirus trifoliata (L.) Raf. into Citrus. J Japan Soc Hort Sci 80:295–307
Oliveria AC, Bastianel M, Christofani-Yaly M, Amaral AM, Machado MA (2007) Development of genetic maps of the citrus varieties ‘Murcott’ tangor and ‘Pê ra’ sweet orange by using fluorescent AFLP markers. J Appl Genet 48:219–231
Ollitrault P, Dambier D, Luro F, Duperray C (1994) Nuclear genome size variation in Citrus. Fruits 49:390–393
Ollitrault P, Terol J, Chen C et al (2012) A reference genetic map of C. clementina hort. ex Tan.; citrus evolution inferences from comparative mapping. 2012. BMC Genomics 13:593
Omura M, Ueda T, Kita M, Komatsu A, Takanokura Y, Shimada T, Endo T, Nesumi H, Yoshida T (2003) EST mapping of Citrus. Proc Int Soc Citricult IX Congr 2000:71–74
Sakamoto T, Miura K, Itoh H et al (2004) An overview of gibberellin metabolism enzyme genes and their related mutants in rice. Plant Physiol 134:1642–1653
Shimada T, Kita M, Endo T, Fujii H, Ueda T, Moriguchi T, Omura M (2003) Expressed sequence tags of ovary tissue cDNA library in Citrus unshiu Marc. Plant Sci 165:167–168
Shimada T, Endo T, Fujii H, Hara M, Ueda T, Kita M, Omura M (2004) Molecular cloning and functional characterization of 4 monoterpene synthase genes from Citrus unshiu Marc. Plant Sci 166:49–58
Siviero AM, Cristofani EL, Furtado A, Garcia FF, Coelho ASG, Machado MA (2006) Identification of QTLs associated with citrus resistance to Phytophthora gummosis. J Appl Genet 47:23–28
Sugiyama A, Ikoma Y, Fujii H, Shimada T, Endo T, Shimizu T, Omura M (2010) Structure and expression levels of alleles of Citrus zeaxanthin epoxidase genes. J Japan Soc Hort Sci 79:263–274
Sugiyama A, Omura M, Matsumoto H, Shimada T, Fujii H, Endo T, Shimizu T, Nesumi H, Ikoma Y (2011) Quantitative trait loci (QTL) analysis of carotenoid content in Citrus fruit. J Japan Soc Hort Sci 80:136–144
Taylor DR, Ingvarsson PK (2003) Common features of segregation distortion in plants and animals. Genetica 117:27–35
Ueda T, Ikeda F, Kita M, Shimada T, Endo T, Omura M (2003) Evaluation of a CAPS method based on ESTs in Citrus. Proc Int Soc Citricult IX Congr 2000:116–117
Vanichanon A, Blake NK, Martin JM, Talbert LE (2000) Properties of sequence-tagged-site primer sets influencing repeatability. Genome 43:47–52
Acknowledgments
This work was supported by a grant from the Ministry of Agriculture, Forestry, and Fisheries of Japan (Genomics-based Technology for Agricultural Improvement, HOR-2003, DNA-marker breeding project).
Data archiving statement
All DNA sequences used in this study were registered in DDBJ and are fully available. Genetic map is now under process of submission to Citrus Genome Database and will be completed during the review.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by W.-W. Guo
Rights and permissions
About this article
Cite this article
Shimada, T., Fujii, H., Endo, T. et al. Construction of a citrus framework genetic map anchored by 708 gene-based markers. Tree Genetics & Genomes 10, 1001–1013 (2014). https://doi.org/10.1007/s11295-014-0738-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11295-014-0738-9