Abstract
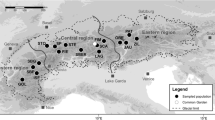
The Mediterranean common shrub Pistacia lentiscus is distributed in a wide range of habitats along the climatic gradient in Israel. We studied the factors that may shape its morphological, physiological, and genetic differentiation. We examined the phenotypic and molecular genetic variability among and within the six Israeli populations as correlated with the local environmental conditions. The genetic structure of the shrub on the island of Cyprus was also examined. Plant morphological parameters correlated significantly with the local environmental conditions, especially with the annual precipitation and temperature. Gene diversity did not differ significantly among locations, and, hence, no differentiation among Israeli populations or between populations in Israel and Cyprus was found. The major part of the molecular variance (69%) was found within the populations, 22% of the variance was found between Israel and Cyprus and 9% among the populations within the region. Gene flow estimates among all the tested populations were high with no indication for the isolation by distance. We did not find any pattern of ecologically related genetic differentiation; hence, the morphological and physiological differences are probably due to phenotypic plasticity. It seems that the ability of P. lentiscus to express the different phenotypes in response to the varying conditions in the Mediterranean region is an adaptive trait in a species that is characterized by intensive gene flow.




Similar content being viewed by others
References
Barazani O, Dudai N, Golan-Goldhirsh A (2003a) Comparison of Mediterranean Pistacia lentiscus genotypes by random amplified polymorphic DNA, chemical, and morphological analyses. J Chem Ecol 29:1939–1952
Barazani O, Atayev A, Yakubov B, Kostiukovsky V, Popov K, Golan-Goldhirsh A (2003b) Genetic variability in Turkmen populations of Pistacia vera L. Genet Resour Crop Evol 50:383–389
Bradshaw AD (1965) Evolutionary significance of phenotypic plasticity in plants. Adv Genet 13:115–155
Bradshaw AD (2006) Unraveling phenotypic plasticity—why should we bother? New Phytol 170:644–648
Braun M (2004) Atlas of Israel. Yavneh Publishing House, Tel Aviv
Coart E, Lamote V, De Loose M, Van Bockstaele E, Lootens P, Roldan-Ruiz I (2002) AFLP markers demonstrate local genetic differentiation between two indigenous oak species [Quercus robur L. and Quercus petraea (Matt.) Liebl] in Flemish populations. Theor Appl Genet 105:431–439
de Jong G (2005) Evolution of phenotypic plasticity: patterns of plasticity and the emergence of ecotypes. New Phytol 166:101–117
Grant MC, Mitton JB (1977) Genetic differentiation among growth forms of Engelmann spruce and subalpine fir at tree line. Arct Alp Res 9:259–263
Gratani L, Meneghini M, Pesoli P, Crescente MF (2003) Structural and functional plasticity of Quercus ilex seedlings of different provenances in Italy. Trees 17:515–521
Hufford KM, Mazer SJ (2003) Plant ecotypes: genetic differentiation in the age of ecological restoration. Trends Ecol Evol 18:147–155
Izhaki I, Safriel UN (1985) Why do fleshy-fruit plants of the Mediterranean scrub intercept fall- but not spring-passage of seed-dispersing migratory birds? Oecologia 67:40–43
Joshi J, Schmid B, Caldeira MC, Dimitrakopoulos PG, Good J, Harris R, Hector A, Huss-Danell K, Jumpponen A, Minns A, Mulder CPH, Pereira JS, Prinz A, Scherer-Lorenzen M, Siamantziouras ASD, Terry AC, Troumbis AY, Lawton JH (2001) Local adaptation enhances performance of common plant species. Ecol Lett 4:536–544
Jump AS, Penuelas J (2005) Running to stand still: adaptation and the response of plants to rapid climate change. Ecol Lett 8:1010–1020
Kandedmir GE, Kandemir I, Kaya Z (2004) Genetic variation in Turkish red pine (Pinus brutia Ten.) seed stands as determined by RAPD markers. Silvae Genet 53:169–175
Kawecki TJ, Ebert D (2004) Conceptual issues in local adaptation. Ecol Lett 7:1225–1241
Kelly CK, Chase MW, de Bruijn A, Fay MF, Woodward FI (2003) Temperature-based population segregation in birch. Ecol Lett 6:87–89
Linhart YB, Grant MC (1996) Evolutionary significance of local genetic differentiation in plants. Annu Rev Ecol Syst 27:237–277
Luikart G, England PR, Tallmon D, Jordan S, Taberlet P (2003) The power and promise of population genomics: from genotyping to genome typing. Nat Rev Genet 4:981–994
Mariette S, Cottrell J, Csaikl UM, Goikoechea P, Konig A, Lowe AJ, Van Dam BC, Barreneche T, Bodenes C, Streiff R, Burg K, Groppe K, Munro RC, Tabbener H, Kremer A (2002) Comparison of levels of genetic diversity detected with AFLP and microsatellite markers within and among mixed Q. petraea (Matt.) Liebl. and Q. robur L. stands. Silvae Genet 51:72–79
Miller MP (1997) Tools for population genetic analyses (TFPGA). Department of Biological Sciences, Northern Arizona University, Flagstaff (AZ)
Mitton JB, Linhart YB, Hamrick JL, Backman J (1977) Population differentiation and mating systems in ponderosa pine in Colorado Front Range. Theor Appl Genet 51:5–14
Nei M (1978) Estimation of average heterozygosity and genetic distance from a small number of individuals. Genetics 89:583–590
Nevo E (2001) Evolution of genome–phenome diversity under environmental stress. Proc Natl Acad Sci USA 98:6233–6240
Nevo E, Beiles A, Ben-Shlomo R (1984) The evolutionary significance of genetic diversity: ecological, demographic and life history correlates. In: Mani GS (ed) Evolutionary dynamics of genetic diversity. Lecture notes in Biomathematics, vol. 53. Springer-Verlag, Berlin, pp 13–213
Peakall R, Smouse PE (2006) GENALEX 6: genetic analysis in Excel. Population genetic software for teaching and research. Mol Ecol Notes 6:288–295
Peng SL, Li QF, Li D, Wang ZF, Wang DP (2003) Genetic diversity of Pinus massoniana revealed by RAPD markers. Silvae Genet 52:60–63
Raymond ML, Rousset F (1995) An exact test for population differentiation. Evolution 49:1280–1283
Scheiner SM (1993) Genetic and evolution of phenotypic plasticity. Annu Rev Ecol Syst 24:35–68
Shaviv I (1978) Autecology of Pistacia Lentiscus L. Doctoral Dissertation. Ph.D. thesis, Technion, Israel Institute of Technology, Haifa, Israel
Slatkin M (1985) Gene flow in natural populations. Annu Rev Ecol Syst 16:393–430
Sultan SE (2005) An emerging focus on plant ecological development. New Phytol 166:1–5
van Kleunen M, Fischer M (2005) Constraints on the evolution of adaptive phenotypic plasticity in plants. New Phytol 166:49–60
Via S, Gomulkiewicz R, De Jong G, Scheiner SM, Peter CD Van Tienderen H (1995) Adaptive phenotypic plasticity: consensus and controversy. Trends Ecol Evol 10:212–217
Vos P, Hogers R, Bleeker M, Reijans M, Van De Lee T, Hornes M, Frijters A, Pot J, Pileman J, Kuiper M, Zabwau M (1995) AFLP: a new technique for DNA fingerprinting. Nuc Acid Res 23:4407–4415
Werner O, Sanchez-Gomez P, Carrion-Vilches MA, Guerra J (2002) Evaluation of genetic diversity in Pistacia lentiscus L. (Anacardiaceae) from the southern Iberian Peninsula and North Africa using RAPD assay. Implications for reafforestation policy. Israel J Plant Sci 50:11–18
West-Eberhard MJ (1989) Phenotypic plasticity and the origins of diversity. Annu Rev Ecol Syst 20:249–278
Yakovlev IA, Kleinschmidt J (2002) Genetic differentiation of pedunculate oak Quercus robur L. in the European part of Russia based on RAPD markers. Russ J Genet 38:148–155
Yeh FC, Yang RC, Boyle TBJ, Ye ZH, Mao JX (1997) POPGENE, the user-friendly shareware for population genetic analysis. Molecular Biology and Biotechnology Centre, University of Alberta
Zohary M (1952) A monographical study of the genus Pistacia. Palest J Bot Jerus Ser 5:187–228
Acknowledgements
We would like to thank A. Shmida for the distribution map of P. lentiscus in Israel.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by A. Abbott
Rights and permissions
About this article
Cite this article
Nahum, S., Inbar, M., Ne’eman, G. et al. Phenotypic plasticity and gene diversity in Pistacia lentiscus L. along environmental gradients in Israel. Tree Genetics & Genomes 4, 777–785 (2008). https://doi.org/10.1007/s11295-008-0150-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11295-008-0150-4