Abstract
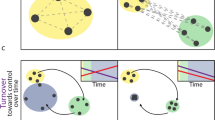
Although the consequences of changes in microbial diversity have received increasing attention, our understanding of processes that drive spatial variation in microbial diversity remains limited. In this study, we sampled bacterial communities in early and late successional temperate forests in Northeast China, and used distance-based redundancy analysis to examine how different processes influence bacterial beta diversity and phylogeny-based beta diversity using the Bray–Curtis and UniFrac metrics, respectively. After controlling for sampling effects, bacterial beta diversity in both forests was higher than expected by chance, which indicates that the bacterial community showed strong intraspecific aggregation. Both environmental filtering and dispersal limitation contributed to bacterial beta diversity and phylogeny-based beta diversity in the two forests. However, the relative importance of these different processes varied between the two forests. In the early successional forest, dispersal limitation played a dominant role in structuring the bacterial community, whereas the effects of environmental filtering were more important in the late successional forest. Our study revealed that bacterial beta diversity and phylogeny-based beta diversity in forest communities from the same region are regulated by different forces and that the relative importance of different forces varies over succession.



Similar content being viewed by others
References
Anderson MJ (2006) Distance-based tests for homogeneity of multivariate dispersions. Biometrics 62:245–253
Bardgett RD, Wardle DA (2010) Aboveground-belowground linkages: biotic interactions, ecosystem processes, and global change. Oxford University, Oxford
Beck S, Powell JR, Drigo B, Gairney JWG, Anderson IC (2015) The role of stochasticity differs in the assembly of soil- and root-associated fungal communities. Soil Biol Biochem 80:18–25
Bezemer TM, Fountain M, Barea J, Christensen S, Dekker S, Duyts H, Van Hal R, Harvey JA, Hedlund K, Maraun M, Mikola J, Mladenov AG, Robin C, De Ruiter P, Scheu S, Setälä H, Šmilauer P, Van der Putten WH (2010) Divergent composition but similar function of soil food webs of individual plants: plant species and community effects. Ecology 91:3027–3036
Borcard D, Legendre P, Avois-Jacquet C, Tuomisto H (2004) Dissecting the spatial structure of ecological data at multiple scales. Ecology 85:1826–1832
Cadotte MW (2007) Concurrent niche and neutral processes in the competition-colonization model of species coexistence. Proc R Soc B 274:2739–2744
Chase JM, Myers JA (2011) Disentangling the importance of ecological niches from stochastic processes across scales. Phil Trans R Soc B 366:2351–2363
Chu H, Fierer N, Lauber CL, Caporaso JG, Knight R, Grogan P (2010) Soil bacterial diversity in the Arctic is fundamentally different from that found in other biomes. Environ Microbiol 12:2998–3006
DeCaceres M, Legendre P, Valencia R, Cao M, Chang L, Chuyong G, Condit R, Hao Z, Hsieh CF, Hubbell S, Kenfack D, Ma K, Mi X, Noor MNS, Kassim AR, Ren H, Su SH, Sun IF, Thomas D, Ye W, He F (2012) The variation of tree beta diversity across a global network of forest plots. Global Ecol Biogeogr 21:1191–1202
Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R (2011) UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27:2194–2200
Ellner SP, Fussmann G (2003) Effects of successional dynamics on metapopulation persistence. Ecology 84:882–889
Ferrenberg S, O’Neill SP, Knelman JE, Todd B, Bradley D, Robinson T, Schmidt SK, Townsend AR, Williams MW, Cleveland CC, Melbourne BA, Jiang L, Nemergut DR (2013) Changes in assembly processes in soil bacterial communities following a wildfire disturbance. ISME J 7:1102–1111
Fierer N, Jackson RB (2006) The diversity and biogeography of soil bacterial communities. Proc Natl Acad Sci USA 103:626–631
Fierer N, Leff JW, Adams BJ, Nielsen UN, Bates ST, Lauber CL, Owens S, Gilbert JA, Wall DH, Caporaso JG (2012) Cross-biome metagenomic analyses of soil microbial communities and their functional attributes. Proc Natl Acad Sci USA 109:21390–21395
Finlay BJ (2002) Global dispersal of free-living microbial eukaryote species. Science 296:1061–1063
Finlay BJ, Clarke KJ (1999) Ubiquitous dispersal of microbial species. Nature 400:828–828
Griffiths RI, Thomson BC, James P, Bell T, Balley M, Whiteley AS (2011) The bacterial biogeography of British soils. Environ Microbiol 13:1642–1654
Kielak A, Pijl AS, van Veen JA, Kowalchuk GA (2008) Differences in vegetation composition and plant species identity lead to only minor changes in soil-borne microbial communities in a former arable field. FEMS Microbiol Ecol 63:372–382
Kowalchuk GA, Buma DS, de Boer W, Klinkhamer PGL, van Veen JA (2002) Effects of above-ground plant species composition and diversity on the diversity of soil-borne microorganisms. Antonie Van Leeuwenhoek 81:509–520
Kraft NJB, Comita LS, Chase JM, Sanders NJ, Swenson NG, Crist TO et al (2011) Disentangling the drivers of b-diversity along latitudinal and elevational gradients. Science 333:1755–1758
Landesman W, Nelson DM, Fitzpatrick MC (2014) Soil properties and tree species drive beta-diversity of soil bacterial communities. Soil Biol Biochem 76:201–209
Legendre P, Mi X, Ren H, Ma K, Yu M, Sun IF, He F (2009) Partitioning beta diversity in a subtropical broad-leaved forest of China. Ecology 90:663–674
Leibold MA, McPeek MA (2006) Coexistence of the niche and neutral perspectives in community ecology. Ecology 87:1399–1410
Li H, Zhang Y, Zhang CG, Chen GX (2005) Effect of petroleum-containing wastewater irrigation on bacterial diversities and enzymatic activities in a paddy soil irrigation area. J Environ Qual 34:1073–1080
Li H, Ye D, Wang X, Settles ML, Wang J, Hao Z, Zhou L, Dong P, Jiang Y, Ma Z (2014) Soil bacterial communities of different natural forest types in Northeast China. Plant Soil 383:203–216
Li H, Wang X, Liang C, Hao Z, Zhou L, Ma S, Li X, Yang S, Yao F, Jiang Y (2015) Aboveground-belowground biodiversity linkages differ in early and late successional temperate forests. Sci Rep 5:12234
Martiny JBH, Eisen JA, Penn K, Allison SD, Horner-Devine MC (2011) Drivers of bacterial beta-diversity depend on spatial scale. Proc Natl Acad Sci USA 108:7850–7854
Monroy F, Van der Putten WH, Yergeau E, Mortimer SR, Duyts H, Bezemer TM (2012) Community patterns of soil bacteria and nematodes in relation to geographic distance. Soil Biol Biochem 45:1–7
Myers JA, Chase JM, Jimenez I, Jorgensen PM, Araujo-Murakami A, Paniagua-Zambrana N, Seidel R (2013) Beta-diversity in temperate and tropical forests reflects dissimilar mechanisms of community assembly. Ecol Lett 16:151–157
Navarrete AA, Tsai SM, Mendes LW, Faust K, de Hollander M, Cassman N, van Veen JA, Kuramae EE (2015) Soil microbiome responses to the short-term effects of Amazonian deforestation. Mol Ecol 24:2433–2448
Nemergut DR, Cleveland CC, Wieder WR, Washenberger CL, Townsend AR (2010) Plot-scale manipulations of organic matter inputs to soils correlate with shifts in microbial community composition in a lowland tropical rain forest. Soil Biol Biochem 42:2153–2160
Nemergut DR, Schmidt SK, Fukami T, O’Neill SP, Bilinski TM, Stanish LF, Knelman JE, Darcy JL, Lynch RC, Wickey P, Ferrenberg S (2013) Patterns and processes of microbial community assembly. Microbiol Mol Biol Rev 77:342–356
Oksanen J, Blanchet FG, Kindt R, Legendre P, Minchin PR, O’Hara RB, Simpson GL, Solymos P, Stevens MHH, Wagner H (2012) Vegan: community ecology package. R package version 2.0–5. http://CRAN.R-project.org/package=vegan
Oliver SG (1996) From DNA sequence to biological function. Nature 379:597–600
Prescott CE, Grayston SJ (2013) Tree species influence on microbial communities in litter and soil: current knowledge and research needs. Forest Ecol Manag 309:19–27
Pruesse E, Quast C, Knittel K, Fuchs BM, Ludwig W, Peplies J, Glöckner FO (2007) SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res 35:7188–7196
R Development Core Team (2012) R: a language and environment for statistical computing. The R Foundation for Statistical Computing, Vienna, Austria. http://www.R-project.org/
Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ, Sahl JW, Stres B, Thallinger GG, Van Horn DJ, Weber CF (2009) Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol 75:7537–7541
Smalla K, Wieland G, Buchner A, Zock A, Parzy J, Kaiser S, Roskot N, Heuer H, Berq G (2001) Bulk and rhizosphere soil bacterial communities studied by denaturing gradient gel electrophoresis: plant-dependent enrichment and seasonal shifts revealed. Appl Environ Microbiol 67:4742–4751
Sokol ER, Herbold CW, Lee CK, Cary SC, Barrett JE (2013) Local and regional influences over soil microbial metacommunities in the Transantarctic Mountains. Ecosphere 4:136
Stephan A, Meyer AH, Schmid B (2000) Plant diversity affects culturable soil bacteria in experimental grassland communities. J Ecol 88:988–998
Wang X, Hao Z, Zhang J, Lian J, Li B, Ye J, Yao X (2009) Tree size distributions in an old-growth temperate forest. Oikos 118:25–36
Wang X, Wiegand T, Hao Z, Li B, Ye J, Lin F (2010) Spatial associations in an old-growth temperate forest, Northeastern China. J Ecol 98:674–686
Yergeau E, Bezemer TM, Hedlund K, Mortlmer SR, Kowalchuk GA, der Putten WH (2010) Influence of space, soil, nematodes and microbial community composition of chalk grassland soils. Environ Microbiol 12:2096–2106
Acknowledgments
This work was supported by the “China Soil Microbiome Initiative: Function and Regulation of Soil-Microbial Systems” of the Chinese Academy of Sciences (XDB15010302), National Natural Science and Foundation of China (31370444), and State Key Laboratory of Forest and Soil Ecology (LFSE2013-11 and LFSE2013-14). T.M. Bezemer was supported by Chinese Academy of Sciences Visiting Professorship for Senior International Scientists (2013T1Z0014). We thank Prof. Pierre Legendre for valuable comments about statistical analyses, Jianjun Wang for helpful comments on the manuscript, our field crew from the Changbai Mountain National Station of Forest Ecosystem Observation and Research for help with collecting the soil cores, and Dr. Suresh Iyer, Dr. Matthew Settles, and the other researchers from the Institute for Bioinformatics and Evolutionary Studies (IBEST), University of Idaho, USA for technical assistance with 454 pyrosequencing analysis.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Electronic supplementary material
Below is the link to the electronic supplementary material.
About this article
Cite this article
Wang, X., Li, H., Bezemer, T.M. et al. Drivers of bacterial beta diversity in two temperate forests. Ecol Res 31, 57–64 (2016). https://doi.org/10.1007/s11284-015-1313-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11284-015-1313-z