Abstract
Objective
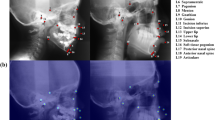
The objective of this work is to present a novel technique using convolutional neural network (CNN) architectures for automatic segmentation of sella turcica (ST) on cephalometric radiographic image dataset. The proposed work suggests possible deep learning approaches to distinguish ST on complex cephalometric radiographs using deep learning techniques.
Materials and methods
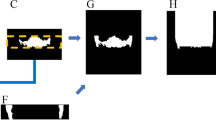
The dataset of 525 lateral cephalometric images was employed and randomly split into different training and testing subset ratios. The ground truth (annotated images) represents pixel-wise annotation of the ST using an online annotation platform by dental specialists. This study compared convolutional neural network architectures based on fine-tuned versions of the VGG19, ResNet34, InceptionV3, and ResNext50 architectures to select an appropriate model for autonomous segmentation of the nonlinear structure of ST.
Results
The study compared training and prediction results of the selected models: VGG19, ResNet34, InceptionV3, and ResNext50. The mean IoU scores for VGG19, ResNet34, InceptionV3 and ResNext50 are 0.7651, 0.7241, 0.4717, 0.4287, dice coefficients are 0.7794, 0.7487, 0.4714, 0.4363 and loss scores are 0.0973, 0.1299, 0.2049 and 0.2251, respectively.
Conclusion
The obtained findings suggest that the VGG19 and Resnet34 architectures (mean IoU and dice coefficient > 75%) comparatively outperformed the InceptionV3 and ResNext50 architectures (mean IoU and dice coefficients is around 45%) for considered cephalometric radiographic dataset. The study findings can be used as a reference model for future investigation of non-linear ST morphological characteristics and related biological anomalies.















Similar content being viewed by others
References
Broadbent BH. A new x-ray technique and its application to orthodontia. Angle Orthod. 1931;1(2):45–66.
Tekiner H, Acer N, Kelestimur F. Sella turcica: an anatomical, endocrinological, and historical perspective. Pituitary. 2015;18(4):575–8.
Svystun O, Schropp L, Wenzel A, Spin-Neto R. Sella turcica area and location of point sella in cephalograms acquired with simulated patient head movements. J Contemp Dent Pract. 2021;22(3):207–14.
Kantor ML, Norton LA. Normal radiographic anatomy and common anomalies seen in cephalometric films. Am J Orthod Dentofacial Orthop. 1987;91(5):414–26.
Subhadra Devi V, Baburao S. Age and sex related morphology and morphometry of sellar region of sphenoid in prenatal and postnatal human cadavers. Int J Res Dev Health. 2013;1(3):141–8.
Costea C, Turliuc S, Cucu A, Dumitrescu G, Carauleanu A, Buzduga C, et al. The “polymorphous” history of a polymorphous skull bone: the sphenoid. Anat Sci Int. 2018;93(1):14–22.
Jones R, Faqir A, Millett D, Moos K, McHugh S. Bridging and dimensions of sella turcica in subjects treated by surgical-orthodontic means or orthodontics only. Angle Orthod. 2005;75(5):714–8.
Muhammed FK, Abdullah AO, Liu Y. Morphology, incidence of bridging, dimensions of sella turcica, and cephalometric standards in three different racial groups. J Craniofac Surg. 2019;30(7):2076–81.
Axelsson S, Storhaug K, Kjær I. Post-natal size and morphology of the sella turcica. Longitudinal cephalometric standards for Norwegians between 6 and 21 years of age. Eur J Orthod. 2004;26(6):597–604.
Alkofide EA. The shape and size of the sella turcica in skeletal Class I, Class II, and Class III Saudi subjects. Eur J Orthod. 2007;29(5):457–63.
Sathyanarayana HP, Kailasam V, Chitharanjan AB. The size and morphology of sella turcica in different skeletal patterns among South Indian population: a lateral cephalometric study. J Indian Orthod Soc. 2013;47(4):266–71.
Shrestha GK, Pokharel PR, Gyawali R, Bhattarai B, Giri J. The morphology and bridging of the sella turcica in adult orthodontic patients. BMC Oral Health. 2018;18(1):1–8.
Zagga A, Ahmed H. Plain radiographic cephalometry of the sella turcica: an overview. Niger J Med. 2008;17(3):333–6.
Björk A, Solow B. Measurement on radiographs. J Dent Res. 1962;41(3):672–83.
Pancherz H. A cephalometric analysis of skeletal and dental changes contributing to Class II correction in activator treatment. Am J Orthod. 1984;85(2):125–34.
Khanagar SB, Al-Ehaideb A, Vishwanathaiah S, Maganur PC, Patil S, Naik S, et al. Scope and performance of artificial intelligence technology in orthodontic diagnosis, treatment planning, and clinical decision-making—a systematic review. J Dent Sci. 2021;16(1):482–92.
Xia J, Qi F, Yuan W, Wang D, Qiu W, Sun Y, et al., editors. Computer aided simulation system for orthognathic surgery. In: Proceedings Eighth IEEE Symposium on Computer-Based Medical Systems; 1995. https://doi.org/10.1109/ICSIGP.1996.567341.
Mosleh MAA, Baba MS, Malek S, Almaktari RA. Ceph-X: development and evaluation of 2D cephalometric system. BMC Bioinform. 2016;17(19):193–201.
Frosio I, Borghese NA, editors. Real time enhancement of cephalometric radiographies. In: 3rd Proc IEEE Int Symp Biomed Imaging: Nano to Macro; 2006. https://doi.org/10.1109/ISBI.2006.1625082.
Mosleh MA, Baba MS, Himazian N, AL-Makramani BM, editors. An image processing system for cephalometric analysis and measurements. In: 2008 International Symposium on Information Technology; 2008. https://doi.org/10.1109/ITSIM.2008.4631953.
Yue W, Yin D, Li C, Wang G, Xu T, editors. Locating large-scale craniofacial feature points on X-ray images for automated cephalometric analysis. In: IEEE International Conference on Image Processing; 2005. https://doi.org/10.1109/ICIP.2005.1530288.
Cardillo J, Sid-Ahmed MA. An image processing system for locating craniofacial landmarks. IEEE Trans Med Imaging. 1994;13(2):275–89.
El-Feghi I, Sid-Ahmed MA, Ahmadi M. Automatic localization of craniofacial landmarks for assisted cephalometry. Pattern Recogn. 2004;37(3):609–21.
Hassan B, van der Stelt P, Sanderink G. Accuracy of three-dimensional measurements obtained from cone beam computed tomography surface-rendered images for cephalometric analysis: influence of patient scanning position. Eur J Orthod. 2009;31(2):129–34.
Swennen GR, Schutyser F. Three-dimensional cephalometry: spiral multi-slice vs cone-beam computed tomography. Am J Orthod Dentofacial Orthop. 2006;130(3):410–6.
Lagravère MO, Carey J, Toogood RW, Major PW. Three-dimensional accuracy of measurements made with software on cone-beam computed tomography images. Am J Orthod Dentofacial Orthop. 2008;134(1):112–6.
Ongkosuwito E, Katsaros C, Van’t Hof M, Bodegom J, Kuijpers-Jagtman A. The reproducibility of cephalometric measurements: a comparison of analogue and digital methods. Eur J Orthod. 2002;24(6):655–65.
Sayinsu K, Isik F, Trakyali G, Arun T. An evaluation of the errors in cephalometric measurements on scanned cephalometric images and conventional tracings. Eur J Orthod. 2007;29(1):105–8.
Zhang L, Luo Z, Chai R, Arefan D, Sumkin J, Wu S, editors. Deep-learning method for tumor segmentation in breast DCE-MRI. In: Medical Imaging 2019: Imaging Informatics for Healthcare, Research, and Applications; 2019: SPIE.
Zhang L, Arefan D, Guo Y, Wu S, editors. Fully automated tumor localization and segmentation in breast DCEMRI using deep learning and kinetic prior. In: Medical Imaging 2020: Imaging Informatics for Healthcare, Research, and Applications; 2020: International Society for Optics and Photonics.
Zhang L, Mohamed AA, Chai R, Guo Y, Zheng B, Wu S. Automated deep learning method for whole-breast segmentation in diffusion-weighted breast MRI. J Magn Reson Imaging. 2020;51(2):635–43.
Celik E, Polat-Ozsoy O, Toygar Memikoglu TU. Comparison of cephalometric measurements with digital versus conventional cephalometric analysis. Eur J Orthod. 2009;31(3):241–6.
Kassani SH, Kassani PH, Wesolowski MJ, Schneider KA, Deters R. Classification of histopathological biopsy images using ensemble of deep learning networks. In: Proceedings of the 29th Annual International Conference on Computer Science and Software Engineering; 2019. p. 92–9.
Kharel N, Alsadoon A, Prasad P, Elchouemi A, editors. Early diagnosis of breast cancer using contrast limited adaptive histogram equalization (CLAHE) and Morphology methods. In: 2017 8th International Conference on Information and Communication Systems (ICICS); 2017. https://doi.org/10.1109/IACS.2017.7921957.
Makandar A, Halalli B. Breast cancer image enhancement using median filter and CLAHE. Int J Sci Eng Res. 2015;6(4):462–5.
Asnaoui KE, Chawki Y, Idri A. Automated methods for detection and classification pneumonia based on x-ray images using deep learning. arXiv preprint http://arxiv.org/abs/200314363. 2020. https://doi.org/10.1007/978-3-030-74575-2_14.
Ronneberger O, Fischer P, Brox T, editors. U-net: Convolutional networks for biomedical image segmentation. In: International Conference on Med Image Comput Comput Assist Interv; 2015. https://doi.org/10.1007/978-3-319-24574-4_28.
LeClerc AJ, Heilenkötter N, Otero Baguer D, Hauberg-Lotte L, Boskamp T, Hetzer S, et al. Deeply supervised UNet for semantic segmentation to assist dermatopathological assessment of basal cell carcinoma. J Imaging. 2021;7(4):71.
Kingma D, Ba J, editors. ADAM: A method for stochastic optimization. In: 3rd International Conference on Learning Representations. Conference Track Proceedings; 2015.
Acknowledgements
The data was provided by the Oral Health Sciences Centre, Postgraduate Institute of Medical Education and Research (PGIMER), Chandigarh.
Funding
Not applicable.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors refuse to have any potential conflicts of interest in the current work.
Ethics approval
Ethical approval was obtained from the Postgraduate Institute of Medical Education and Research (PGIMER), Chandigarh Institutional Ethics Committee (IEC-09/2021-2119). Furthermore, this study was conducted with approval no: PGI/IEC/2021/001488. ‘This article does not contain any studies with human or animal subjects performed by the any of the authors.’
Informed consent
Not applicable.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Shakya, K.S., Laddi, A. & Jaiswal, M. Automated methods for sella turcica segmentation on cephalometric radiographic data using deep learning (CNN) techniques. Oral Radiol 39, 248–265 (2023). https://doi.org/10.1007/s11282-022-00629-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11282-022-00629-8