Abstract
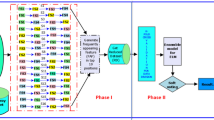
Microarray-based gene expression profiling is an emerging method to predict, classify, diagnose and to treat cancer efficiently. The characteristics of this cancer disease may change frequently which creates large volume of data. In this paper we propose a Novel Map reduced based parallel feature selection and extreme learning for micro array cancer data classification. Initially the gene expression data sets are pre-processed by attribute-wise normalization and setting thresholds on the original data. The second phase uses a wrapper model that uses Adaptive Whale Optimization Algorithm (AWOA) with Nelder–Mead algorithm (NMA) to accomplish the feature (gene) subset selection. Wrapper models are used to describe the selection process of feature sets as a search issue. Here, various combinations are formulated, estimated and compared with other combinations. At last, to demonstrate the effectiveness of the selected genes using the proposed feature selection method, a Regularized Extreme Learning Machine (RELM) classifier is used to classify the gene expression data subsets chosen by AWOA algorithm.











Similar content being viewed by others
Availability of data and material
Data sharing not applicable to this article as no datasets were generated or analysed during the current study.
Code availability
Custom code.
References
Leung, Y. F., & Cavalieri, D. (2003). Fundamentals of cDNA microarray data analysis. Trends in Genetics, 19(11), 649–659.
Kumar, M., Rath, N. K., & Rath, S. K. (2016). Analysis of microarray leukemia data using an efficient MapReduce-based K-nearest-neighbor classifier. Journal of Biomedical Informatics., 60, 395–409.
Golub, T. R., Slonim, D. K., Tamayo, P., Huard, C., Gaasenbeek, M., Mesirov, J. P., Coller, H., Loh, M. L., Downing, J. R., & Caligiuri, M. A. (1999). Molecular classification of cancer: Class discovery and class prediction by gene expression monitoring. Science, 286(5439), 531–537.
Kumar, M., & Rath, S. K. (2015). Classification of microarray using MapReduce based proximal support vector machine classifier. Knowledge-Based Systems., 89, 584–602.
Hernandez, J. C. H., Duval, B., & Hao, J.-K. (2007). A genetic embedded approach for gene selection and classification of microarray data. Evolutionary Computation (pp. 90–101). Springer.
Peng, Y., Li, W., & Liu, Y. (2006). A hybrid approach for biomarker discovery from microarray gene expression data for cancer classification. Cancer Informatics, 2(301).
Youcong, N., Zhiqiang, Y., & Ruliang, X. (2013). High performance parallel evolutionary algorithm model based on MapReduce framework. International Journal of Computer Application Technology., 46(3), 290–295.
Chen, A. H., & Lin, C. H. (2011). A novel support vector sampling technique to improve classification accuracy and to identify key genes of leukaemia and prostate cancers. Expert Systems with Applications, 38(4), 3209–3219.
Pradipta, M., & Chandra, D. (2012). Relevant and significant supervised gene clusters for Microarray cancer classification. NanoBioscience., 11(2), 161–168.
Kdogan, A., Demiryurek, U., & Banaei-Kashani, F. S. (2010). Voronoi-based geospatial query processing with map reduce. In: Cloud computing technology and science (CloudCom), 2nd international conference on IEEE (pp. 9–16).
Schatz, M. C. (2009). Cloud burst: Highly sensitive read mapping with MapReduce. Bioinformatics, 25(11), 1363–1369.
Ding, W., Lin, C.-T., & Chen, S. (2018). Multiagent-consensus-MapReduce-based attribute reduction using co-evolutionary quantum PSO for big data applications. Neurocomputing, 272, 136–153.
Cho, J.-H., Lee, D., Park, J. H., & Lee, I.-B. (2004). Gene selection and classification from microarray data using kernel machine. Elsevier., 571(1), 93–98.
Caruana, G., Li, M., & Qi, M. A. (2011). MapReduce based parallel SVM for large scale spam filtering Fuzzy systems and knowledge discovery (FSKD). In 2011 8th international conference (Vol. 4, pp. 2659–2662)
Kiran, M., Kumar, A., & Mukherjee, S. P. (2013). Verification and validation of MapReduce program model for parallel support vector machine algorithm on Hadoop. Cluster, 10, 317–325.
Low, Y., Gonzalez, J., Kyrola, A., Bickson, D., Guestrin, C., & Hellerstein, J. M. (2010). Graphlab a new parallel framework for machine learning. Conference on uncertainty in artificial intelligence (UAI)
Ghesmoune, M., Lebbah, M., & Azzag, H. (2015). Micro-batching growing neural gas for clustering data streams using spark streaming. Procedia Computer Science INNS Conference on Big Data 2015Program San Francisco, 53, 158–166
Karau, H., Konwinski, A., Wendell, P., & Zaharia M. (2015). Learning spark: Lightning-fast big data analytics. O’Reilly Media, Incorporated.
Hosseini, B., & Kiani, K. (2018). FWCMR: A scalable and robust fuzzy weighted clustering based on MapReduce with application to microarray gene expression. Expert Systems with Applications., 91, 198–210.
Yan, X., Zhu, Z., & Wu, Q. (2018). Intelligent inversion method for pre-stack seismic big data based on MapReduce. Computers & Geosciences, 110, 81–89.
Chu, C.-T., Kim S., Lin, Y.-A., Yu, Y., Bradski, G., Ng, A., & Olukotun, K. (2007). Map-reduce for machine learning on multicore. Advances in Neural Information Processing Systems, 281–288.
Boeva, V. (2014). Clustering approaches for dealing with multiple DNA microarray datasets. Journal of Computational Science., 5(3), 368–376.
Kumar, M., Rath, N. K., & Swain, A. (2015). Feature selection and classification of microarray data using MapReduce based ANOVA and K-nearest neighbor. Procedia Computer Science, 54, 301–310.
Islam, T., Jeong, B.-S., & Bari, G. (2015). MapReduce based parallel gene selection method. ApplIntell, 42, 147–156.
Mennour, R., & Batouche, M. (2015). Drug discovery for breast cancer based on big data analytics techniques. International Conference on Information & Communication Technology and Accessibility (ICTA), 1–6.
Jenifer, X. R., & Lawrance, R. (2016). An adaptive classification model form microarray analysis using big data. International Conference on Computing Technologies and Intelligent Data Engineering (ICCTIDE'16), 1–5.
Alshamlan, H., Badr, G., & Alohali, Y. (2015). mRMR-ABC: A hybrid gene selection algorithm for cancer classification using microarray gene expression profiling. BioMed Research International.
Alshamlan, H. M., Badr, G. H., & Alohali, Y. A. (2015). Genetic bee colony (GBC) algorithm: A new gene selection method for microarray cancer classification. Computational Biology and Chemistry, 56, 49–60.
Lu, H., Chen, J., Yan, K., Jin, Q., Xue, Y., & Gao, Z. (2017). A hybrid feature selection algorithm for gene expression data classification. Neurocomputing, 256, 56–62.
Salem, H., Attiya, G., & El-Fishawy, N. (2017). Classification of human cancer diseases by gene expression profiles. Applied Soft Computing, 50, 124–134.
Aziz, R., Verma, C. K., & Srivastava, N. (2017). A novel approach for dimension reduction of microarray. Computational Biology and Chemistry, 71, 161–169.
Moradi, P., & Gholampour, M. (2016). A hybrid particle swarm optimization for feature subset selection by integrating a novel local search strategy. Applied Soft Computing, 43, 117–130.
Dashtban, M., & Balafar, M. (2017). Gene selection for microarray cancer classification using a new evolutionary method employing artificial intelligence concepts. Genomics, 109(2), 91–107.
Dashtban, M., Balafar, M., & Suravajhala, P. (2018). Gene selection for tumor classification using a novel bio-inspired multi-objective approach. Genomics, 110(1), 10–17.
Sharbaf, F. V., Mosafer, S., & Moattar, M. H. (2016). A hybrid gene selection approach for microarray data classification using cellular learning automata and ant colony optimization. Genomics, 107(6), 231–238.
Vural, H., & Subaşı, A. (2015). Data-mining techniques to classify microarray gene expression data using gene selection by SVD and information gain. Modeling of Artificial Intelligence, 2, 171–182.
Kar, S., Sharma, K. D., & Maitra, M. (2015). Gene selection from microarray gene expression data for classification of cancer subgroups employing PSO and adaptive K-nearest neighborhood technique. Expert Systems with Applications, 42(1), 612–627.
Mirjalili, S., & Lewis, A. (2016). The whale optimization algorithm. Advances in Engineering Software., 95, 51–67.
Huang, G.-B., Zhou, H., Ding, X., & Zhang, R. (2011). Extreme learning machine for regression and multiclass classification. IEEE Transactions on Systems, Man, and Cybernetics, Part B (Cybernetics), 42(2), 513–529.
Maronna, R. A. (2011). Robust ridge regression for high-dimensional data. Technometrics, 53(1), 44–53.
Motieghader, H., Najafi, A., Sadeghi, B., & Masoudi-Nejad, A. (2017). A hybrid gene selection algorithm for microarray cancer classification using genetic algorithm and learning automata. Informatics in Medicine Unlocked, 9, 246–254.
Funding
None.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
Swati Hira and Anita Bai declared that there is no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Hira, S., Bai, A. A Novel Map Reduced Based Parallel Feature Selection and Extreme Learning for Micro Array Cancer Data Classification. Wireless Pers Commun 123, 1483–1505 (2022). https://doi.org/10.1007/s11277-021-09196-3
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11277-021-09196-3