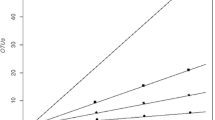
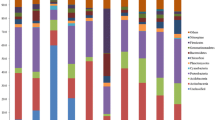
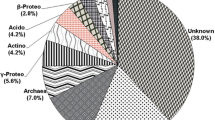
Abstract
The bacterial diversity in the forest soil of Kashmir, India was investigated by 16S rDNA-dependent molecular phylogeny. Small subunit rRNA (16S rDNA) from forest soil metagenome were amplified by polymerase chain reaction (PCR) using primers specific to the domain bacteria. 30 unique phylotypes were obtained by PCR based RFLP of 16S rRNA genes using endonucleases Hae 111 and Msp 1, which were most suitable to score the genetic diversity. The use of 16S rRNA analysis allowed identification of several bacterial populations in the soil belonging to the following phyla: Firmicutes (33.3%), Bacteroidetes (13.3%), Proteobacterium (6.6%), Planctomycete (3.3%), and Deferribacteraceae (3.3%) in addition to the others that were not classified, beyond Archaea domain, However, 36.6% of the retrieved bacterial sequences could not be grouped with any phylum/lineage. The large amount of unclassified clone sequence could imply that novel groups of bacteria were present in the forest soil.

Similar content being viewed by others
References
Ahmad N, Sharma S, Khan FG, Kumar R, Johri S, Abdin MZ, Qazi GN (2008) Phylogenetic analyses of archaeal ribosomal DNA Sequences from Salt Pan Sediment of Mumbai, India. Curr Microbiol 57:145–152. doi:10.1007/s00284-008-9167-z
Amann RI, Ludwig W, Schleifer KH (1995) Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol J Rev 59:143–169
Axelrood PE, Chow ML, Radomski CC, McDermott JM, Davies J (2002) Molecular characterization of bacterial diversity from British Columbia forest soils subjected to disturbance. Can J Microbiol 48:655–674. doi:10.1139/w02-059
Bernardet JF, Vancanneyt M, Matte-Tailliez O, Grisez L, Tailliez P, Bizet C, Nowakowski M, Kerouault B, Swings J (2005) Polyphasic study of Chryseobacterium strains isolated from diseased aquatic animals. Syst Appl Microbiol 28:640–660
Bissett A, Bowman J, Burke C (2006) Bacterial diversity in organically-enriched fish farm sediments. FEMS Microbiol Ecol 55:48–56. doi:10.1111/j.1574-6941.2005.00012.x
Borneman J, Triplett EW (1997) Molecular microbial diversity in soils from eastern Amazonia: evidence for unusual microorganisms and microbial population shifts associated with deforestation. Appl Environ Microbiol 63:2647–2653
Chow ML, Radomski CC, McDermott JM, Davies J, Axelrood PE (2002) Molecular characterization of bacterial diversity in Lodgepole pine (Pinus contorta) rhizosphere soils from British Columbia forest soils differing in disturbance and geographic source. FEMS Microbiol Ecol 42:347–357. doi:10.1111/j.1574-6941.2002.tb01024.x
Dunbar JL, Ticknor O, Kuske CR (2000) Assessment of microbial diversity in four southwestern United States soils by 16S rRNA gene terminal restriction fragment analysis. Appl Environ Microbiol 66:2943–2950. doi:10.1128/AEM.66.7.2943-2950.2000
Griepenburg U, Ward-Rainey N, Mohamed S, Schlesner H, Marxsen H, Rainey FA et al (1999) Phylogenetic diversity of polyamine pattern and DNA base composition of members of the order Planctomycetales. Int J Syst Bacteriol 49:689–696
Gurtler V, Stanisich VA (1996) New approaches to typing and identification of bacteria using the 16S–23S rDNA spacer region. Microbiology 142:3–16
Hackl E, Zechmeister-Boltenstern S, Bodrossy L, Sessitsch A (2004) Comparison of diversities and compositions of bacterial populations inhabiting natural forest soils. Appl Environ Microbiol 70:5057–5065. doi:10.1128/AEM.70.9.5057-5065.2004
Hugenholtz P, Huber T (2003) Chimeric 16S rDNA sequences of diverse origin are accumulating in the public databases. Int J Syst Evol Microbiol 53:289–293. doi:10.1099/ijs.0.02441-0
Komatsoulis GA, Waterman MS (1997) A new computational method for detection of chimeric 16S rRNA artifacts generated by PCR amplification from mixed bacterial populations. Appl Environ Microbiol 63:2338–2346
Kopczynski ED, Batson MM, Ward DM (1994) Recognition of chimeric small-subunit ribosomal DNAs composed of genes from uncultivated microorganisms. Appl Environ Microbiol 60:746–748
Oline DK, Schmidt SK, Grant MC (2006) Biogeography and landscape-scale diversity of the dominant Crenarchaeota of soil. Microb Ecol 52:480–490. doi:10.1007/s00248-006-9101-5
Olsen GJ, Lane DJ, Giovannoni SJ, Pace NR, Stahl DA (1986) Microbial ecology and evolution: a ribosomal RNA approach. Annu Rev Microbiol 40:337–365. doi:10.1146/annurev.mi.40.100186.002005
Pace NR (1997) A molecular view of microbial diversity and the biosphere. Science 276:734–740. doi:10.1126/science.276.5313.734
Park SJ, Yoon JC, Shin KS, Kim EH, Yim S, Cho YJ, Sung GM, Lee DG, Kim SB, Lee DU, Woo SH, Koopman B (2007) Dominance of endospore-forming bacteria on a Rotating Activated Bacillus Contactor biofilm for advanced wastewater treatment. J Microbiol 45:113–121
Raina AN (2002) Geography of Jammu and Kashmir state. Radha Krishan Anand & Co., Pacca Danga, Jammu
Shuldiner AR, Nirula A, Roth J (1989) Hybrid DNA artifact from PCR of closely related target sequences. Nucleic Acids Res 17:4409. doi:10.1093/nar/17.11.4409
Tarlera S, Jangid K, Ivester AH, Whitman WB, Williams MA (2008) Microbial community succession and bacterial diversity in soils during 77, 000 years of ecosystem development. FEMS Microbiol Ecol 64:129–140
Upchurch R, Chiu CY, Everett K, Dyszynski G, Coleman DC, Whitman WB (2008) Differences in the composition and diversity of bacterial communities from agricultural and forest soils. Soil Biol Biochem 40:1294–1305. doi:10.1016/j.soilbio.2007.06.027
Ward DM, Weller R, Bateson MM (1990) 16SrRNA sequences reveal numerous uncultured microorganisms in a natural community. Nature 345:63–65. doi:10.1038/345063a0
Weisburg WG, Barns SM, Pelletier DA, Lane DJ (1991) 16S ribosomal DNA amplification for phylogenetic study. J Bacteriol 173:697–703
Acknowledgments
Nasier Ahmad is thankful to Council of Scientific and Industrial Research (CSIR), Govt. of India for the award of Senior Research fellowship. The authors are grateful to Rajinder Kumar for excellent technical assistance in sequencing work and Dr. Syed Riyaz ul Hassan for providing the soil samples and helpful suggestions. This work was financially supported by CSIR, New Delhi, project Number NWP006.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Ahmad, N., Johri, S., Abdin, M.Z. et al. Molecular characterization of bacterial population in the forest soil of Kashmir, India. World J Microbiol Biotechnol 25, 107–113 (2009). https://doi.org/10.1007/s11274-008-9868-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11274-008-9868-2