Abstract
This study was aimed for the detection of Vibrio parahaemolyticus by biochemical and molecular methods in seafood samples collected from the markets of Cochin located at the southwest coast of India. A total of seventy-two V. parahaemolyticus cultures were isolated by selecting sucrose and cellobiose non-fermenting colonies. All the biochemically confirmed strains were found to have 368-bp toxR gene fragment, while an additional 24% of the samples were confirmed as V. parahaemolyticus by toxR based polymerase chain reaction (PCR) from enrichment broths. PCR based methods are used to detect tdh, trh, and orf8 genes for the identification of pathogenic and pandemic V. parahaemolyticus. Only one out of two urease positive isolates amplified the trh (500bp) gene. About 10% of the isolates showed weak haemolysis and none were found to amplify tdh (269 bp) and orf8 (746 bp) genes, thus indicating the meager incidence of pandemic strains from this area. The incidence of trh positive isolates from market samples signals towards the adoption of stringent seafood safety measures for the products meant for human consumption.
Similar content being viewed by others
Introduction
Vibrio parahaemolyticus is a prevalent seafood-borne pathogen in many Asian countries where marine foods are frequently consumed (Joseph et al. 1983). The organism has been reported as the causative agent of gastroenteritis (Fujino et al. 1953), wound infections, and septicemia because of the consumption of contaminated seafoods. Recently with the emergence of pandemic O3:K6 strain of V. parahaemolyticus that was reported to cause acute gastroenteritis, this pathogen has acquired greater significance (Matsumoto et al. 2000). Epidemiological studies revealed an association between the Kanagawa positive phenomenon (KP+) and gastroenteritis (Okuda et al. 1997a). In the 1980s few gastroenteritis cases were reported from the KP negative V. parahaemolyticus isolates, which led to the discovery of the TDH-related haemolysin (TRH) (Hervio-Heath et al. 2002). Studies revealed the thermostable direct haemolysin (TDH) and the TDH-related haemolysin (TRH), encoded by the tdh and trh genes, respectively, as the major virulence factors of this organism (Shirai et al. 1990). The test adopted for Kanagawa phenomenon using Wagatsuma agar consumes time and labor. Moreover, the test may not be determinative as many environmental strains are Kanagawa negative and do not produce TDH (Tada et al. 1992). Presently there is no in-vitro test for the detection of TRH in V. parahaemolyticus. Sensitive and rapid molecular method such as PCR has been applied to identify the presence of tdh and trh genes from V. parahaemolyticus (Wong et al. 1999; Kim et al. 1999).
A unique clone of V. parahaemolyticus O3:K6 was responsible for many of the recent V. parahaemolyticus outbreaks, reported in India, Russia, Southeast Asia, Japan, and North America (Matsumoto et al. 2000). During 1996 the strains of the O3:K6 serovar was reported to emerge in the Eastern province of India accounting for about 50–80% of V. parahaemolyticus infections annually. It was determined that a filamentous phage, f237 is specifically and exclusively associated with the O3:K6 serovar (Nasu et al. 2000), which was found to have 10 open reading frames (ORFs), including a unique open reading frame, ORF8. Vibrio cholerae has a similar filamentous phage CTX, which carries the cholera enterotoxin genes ctxA and ctxB (Iida et al. 2001). In V. parahaemolyticus O3:K6 serotype, the f237 phage possesses orf8 in place of ctxAB (Waldor and Mekalanos 1996). It was therefore suggested that orf8 gene might be associated with the virulence characteristics of the strains (Okuda et al. 1997b). Nasu et al. (2000) isolated a filamentous phage possessing orf8 in another recently emerging serovar (O4:K68), and reported the orf8 gene fragment as a useful genetic marker for the pandemic group. A toxin regulatory gene (toxR) sequence specific to V. parahaemolyticus, which is present in all the strains irrespective of their ability to produce TDH or TRH, has been applied for definitive identification of the bacterial isolates by several workers (Jacksic et al. 2002; Tada et al. 1992; Kim et al. 1999).
Cochin is a major fish-landing centre in south west coast of India, and accounts for over 90% of statewide exports. Earlier studies carried out at Cochin to identify virulent strains of V. parahaemolyticus, were based on the traditional methods using phenotypic assays and biochemical tests, which require several days for confirmation (Sanjeev and Stephen 1992). In this study an attempt has been made to determine the occurrence of V. parahaemolyticus in seafoods marketed in this area, and to detect the presence of virulent isolates carrying tdh, trh, and orf8 genes from sample isolates and their enrichment broths by biochemical and PCR methods.
Materials and methods
Sample collection and processing
A total of 12 finfish, 25 shellfish, and 5 cephalopod samples were collected over a period of one year from various retail fish markets in and around Cochin for microbiological examination. Finfish samples included Oreochromis mossambicus, Mugil cephalus, Sardinella longipes, Etroplus suratensis, Pampus argentius, Megalaspis cordyla, and Labeo rohita. Shellfishes consisted of Penaeus monodon, Fenneropenaeus indicus, Parapenaeopsis stylifera, Panulirus homarus, Metapenaeus dobsoni, Metapenaeus affinis, Macrobrachium rosenbergii, and Scylla serrata. Cephalopod samples included Sepia pharonis and Loligo duvacelli. All the samples were collected in sterile containers or sterile polythene bags and processed within 2 h of their collection. In brief, heads and tails of the fish were cut off, and the guts were removed. In the case of shrimp, whole mass was taken and cut into small pieces with sterile scissors, while in case of cephalopods; the meat was picked from different parts.
Enrichment, isolation, and identification of V. parahaemolyticus
V. parahaemolyticus was isolated and identified as described in the Bacteriological Analytical Manual of the Food and Drug Administration (Elliot et al. 1995) with modifications. Briefly, the process of identification was initiated with the selection of sucrose followed by cellobiose non-fermenting colonies, and further proceeded with other biochemical tests unlike the reported method (Elliot et al. 1995).
The samples (25 g) were homogenized with alkaline peptone water (APW) broth (225 ml) in a sterile polythene stomacher bag (Stomacher 400 Seaward medicals, UK) at 230 rpm for 1 min, and enriched in APW broth for 18–24 h. Following 8 h of incubation, samples from enrichment broth (1 ml) was drawn aseptically into a microfuge tube for PCR experiment. After 18–24 h of incubation the broth (0.5 ml) was aseptically pipetted into thiosulphate citrate bilesalt sucrose agar (TCBS) plates previously preset and dried (56°C, 45 min). The plates were incubated at 37°C for 24–30 h. About 3–4 typical colonies having green or bluish green color with dark blue or green centre measuring about 3–5 mm were picked from TCBS plate, and each one was inoculated into sterile sucrose broth followed by cellobiose medium supplemented with NaCl (3% w/v). Sucrose and cellobiose non-fermenting colonies were streaked onto sterile tryptone soy agar slants (TSA) supplemented with NaCl (3% w/v) and maintained at room temperature for further identification. Halophilism tests were performed using tryptone broth with different concentrations of NaCl (0, 3, 6, 8, and 10% w/v). Additional characterization tests for the identification of V. parahaemolyticus namely, Gram staining, catalase, cytochrome oxidase tests, triple sugar iron tests, lysine iron agar tests, arginine dehydrolase tests, lysine and ornithine decarboxylase test, and O/129 susceptibility tests were performed (FDA 1992). Tests for glucose oxidation-fermentation were carried out using Hugh-Leifson broth, and arabinose, lactose, mannitol, mannose, salicin and inositol fermentation tests were also performed. All the media were supplemented with NaCl (3% w/v) unless otherwise specified. Colonies that gave typical colony characteristics were picked up and confirmed by Kanagawa (Wagatsuma 1968) and urease tests (Andrews and Hammack 2001).
The isolates exhibiting atypical reaction for any one or two of the sugars or amino acids were also maintained for further analyses. The presumed positive cultures were confirmed using RAPID Hi-Vibrio™ identification kit (KB007, Himedia, India) containing twelve different dehydrated medias (Voges Proskauer, Arginine, ONGP, citrate, ornithine, mannitol, arabinose, sucrose, glucose, salicin, cellobiose and salt concentration) in a single strip. In brief, individual colonies of V. parahaemolyticus grown on nutrient agar (3% w/v, NaCl) were isolated and inoculated in alkaline peptone water (APW, 5 ml) supplemented with NaCl (3% w/v). The broth was incubated at 37°C for 18–24 h until the turbidity was found to be greater than or equal to 0.5 OD at 620 nm. Each well, of the strip containing dehydrated media was aseptically inoculated with 50 μl of the inoculum by surface inoculation method, and the strip was incubated at 35–37°C for 18–24 h. Results were interpreted following standards given in the identification index supplied with the kit.
Haemolytic activity
The Kanagawa phenomenon (KP) or TDH phenotype detection was carried out on Wagatsuma agar (Wagatsuma 1968) as described by Elliot et al. (1992). Haemolytic activity was determined following the procedures of Kishishita et al. (1992). In brief V. parahaemolyticus strains were seeded in Wagatsuma agar by stab and surface inoculation methods, incubated at 37°C for 20–24 h. Isolates producing a clear zone (β-haemolysis) around the colonies were identified as Kanagawa positives while those with weak haemolysis (α–haemolysis) and no hemolytic zone (γ–haemolysis) were termed as Kanagawa negative strains.
Determination of urease activity
Urease activity was determined as described by Andrews and Hammack (2001) with modifications. Briefly, urea agar slants were prepared with an additional incorporation of NaCl (1% w/v) to the Christensen’s urea agar (1 g of peptone, 20 g of NaCl, 2 g of KH2PO4, 1 g of dextrose, 15 g of agar, and phenol red 0.012 g in 1 l of distilled water; pH 6.8). Urea solution (0.5 ml of 40% w/v) was added to the sterile agar medium under aseptic conditions and allowed to cool to form slants. In order to characterize the TRH phenotype, bacterial cultures were streak inoculated and kept for incubation at 36 ± 1°C for 24–48 h. Development of pink color in the urea agar slant was indicated as positive reaction, while the negative cultures retained the media color (yellow).
Preparation of bacterial lysate and PCR assay
Bacterial lysate was prepared following established procedure (Ausubel et al. 1987) with modifications. The biochemically identified isolate was streaked on TSA plates and the colonies were inoculated in the Luria Bertane (LB) broth supplemented with NaCl (2% w/v), and incubated at 37°C under shaking (120 rpm) for 16–18 h. The broth cultures were centrifuged (10,000 rpm, 4°C, 1 min; 5804 R, Eppendorf, Germany) to obtain the pellet, which was washed with normal saline (0.8% NaCl w/v) and re-suspended with DNA-free sterile distilled water (200 μl). The resulting suspension was heated at 98 ± 2°C for 15–20 min in a water bath to lyse the cells, and release the DNA. The lysate was clarified by centrifugation (10,000 rpm, 4°C, 5 min), and the supernatant was stored (−20°C) until further use.
The PCR targeting toxR, tdh, trh, and orf8 genes of V. parahaemolyticus were performed separately both from the characterized isolates and the sample enrichment broths as described previously (Bej et al. 1999; Kim et al. 1999, Nasu et al. 2000) using a thermocycler (Eppendorf Mastercycler, Germany).
PCR amplification was optimized in a total reaction volume of 25 μl consisting of sterile Milli Q water (13.5 μl), 10× PCR buffer (2.5 μl), primer (1.5 μl), dNTP mix (0.5 μl, 200 mM), template (5 μl), and Taq DNA polymerase (0.5 μl). The components were mixed well and the PCR amplification of the target sequence was arranged in a thermocycler (Eppendorf Mastercycler, Germany), and programmed for 30 cycles of amplification. Each cycle consisted of three step reactions i.e., initial denaturation (94°C, 3 min) followed by 30 cycles of denaturation (94°C, 1 min), and extension (72°C, 1 min) followed by final extension (72°C, 5 min). Variation was found in the annealing temperatures for each gene as 60°C for toxR, 58°C for trh, 56°C for tdh, and 57°C for orf8 for 1 min.
The PCR products were resolved on agarose (1.5% w/v) gel electrophoresis. The gel was stained with ethidium bromide (0.5 mg/ml) and visualized under a UV transilluminator (Alpha Imager, Innotech Corporation, USA). GeneRuler™ 100 bp DNA Ladder (MBI Fermentas, USA) was used as a molecular size marker.
Primers
The primers used in this study to detect the targeted genes namely, toxR (toxin regulatory gene), tdh (thermostable direct hemolysin gene), trh (tdh-related hemolysin gene), and orf8 (open reading frame gene) are referred in Table 1.
DNA sequencing
The primers based on the toxR and trh gene fragments were used to amplify and sequence DNA of these amplicons using Beckman coulter CEQ 8000 genetic analysis system (Bioserve Biotechnologies, India). V. parahaemolyticus isolates, which generated amplicons of the expected size i.e., 500 bp for trh and 368 bp for toxR were used for sequencing. Sequence assembly, analysis, and alignments were performed with DNA using Chromas LITE software (Version 2.01, Technelysium Pvt. Ltd., Australia). The nucleotide sequence data was analyzed for homology with the published sequence in the GenBank database using the Basic Local Alignment Search Tool (BLAST), and percent homology was noted. The nucleotide sequences were submitted to the GenBank to obtain the accession numbers. The accession numbers obtained from the GenBank for the selected isolates were bankit914935 EF640377 and bankit843662 DQ997814 for toxR and trh gene sequences, respectively.
Results
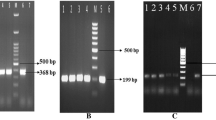
The incidences of V. parahaemolyticus were found to be 41.6, 64, and 40% by conventional method in finfish, shellfish, and cephalopods, respectively. The isolates which were initially identified, as V. parahaemolyticus by conventional biochemical tests were further confirmed by PCR targeting species specific toxR (368 bp) gene. An additional 25%, 20%, and 40% were noticed in the sample enrichment broths by PCR targeting toxR gene in finfish, shellfish, and cephalopods, respectively. Seventy-two of the isolates, which were confirmed to be V. parahaemolyticus by RAPID Hi-Vibrio™ identification kit were further confirmed by PCR targeting species specific toxR (368 bp) gene. All the isolates were found to amplify toxR gene by PCR analysis thus confirming the identity of the bacteria (Fig. 1). By conventional method twenty-three out of 42 samples (54.8%) were found to be positive for V. parahaemolyticus, while an additional 24% of the samples were confirmed for the same with toxR based PCR method using enrichment broths (Table 2).
Agarose gel electrophoresis of PCR products amplified for toxR (368 bp) gene. Lane M: DNA ladder (100 bp); Lane 1: MTCC 451 (type strain); Lanes 2–7: biochemically characterized V. parahaemolyticus isolates; Lanes 8–13: enrichment broths from the samples showing atypical V. parahaemolyticus isolates, Lane 14: PCR negative control
The suspected V. parahaemolyticus isolates were identified by a battery of biochemical tests following the method listed in Bacteriological Analytical Manual (FDA 1992). However, certain isolates were reported to exhibit variation in one or two biochemical reactions (Dileep et al. 2003). Such atypical reactions may pose difficulties in identification of the organism (Karunasagar et al. 1997). The identification method based on the toxR-targeted PCR was developed to solve this problem (Kim et al. 1999). In this study, the isolates were tested for the fermentation of six different sugars and three amino acids. The isolates that exhibited variations in the utilization of one or two sugars or amino acids were considered as atypical isolates. A total of nine atypical isolates were recovered of which five were confirmed to be V. parahaemolyticus by the PCR targeting toxR gene.
In this study, 2 out of 72 (2.8%) isolates showed urease activity of which only one (1.4%) isolate, obtained from Metapenaeus dobsoni sample exhibited amplification for trh gene (Fig. 2). The other urease positive strain, which did not amplify trh gene, was isolated from another inshore shrimp, M. affinis. The trh gene was not amplified from the urease positive isolate of M. affinis was evident from the PCR of the biochemically confirmed strains and the sample enrichment broths. Although 10% of the 72 V. parahaemolyticus isolates showed weak haemolysis, none of them were found to exhibit positive amplification for the tdh (269 bp) gene or β-haemolysis on Wagatsuma agar. Though toxR gene was amplified from all the isolates, none of these strains exhibited positive amplification for orf8 gene.
Agarose gel electrophoresis of PCR products amplified for trh (500 bp) gene. Lane M: DNA ladder (100 bp); Lane 1–3: isolates negative for the gene; Lane 4: type strain (NCMB 1902) positive for trh gene; Lane 5: isolate from M. dobsoni sample containing trh gene; Lane 6: isolate from M. affinis sample without trh gene, showing urease activity; Lane 7: PCR negative control
The nucleotide sequences of the amplified products were analyzed for homology with the published sequence in the GenBank database using the Basic Local Alignment Search Tool (BLAST) and percent homology was noted. The toxR and trh gene fragments exhibited 95–97% and 93–97% similarity, respectively with the published sequences.
Discussion
The members of the genus Vibrio have been frequently defined as opportunistic and potential pathogenic bacteria of the tropical waters (Huss 1997). In India the incidence of V. parahaemolyticus causing food poisoning due to the consumption of contaminated seafoods was reported to be doubled in the last 5 years (Choudhury et al. 2000). Therefore detection of this organism in seafoods is very important.
In this study the detection of V. parahaemolyticus was carried out by conventional biochemical and modern PCR methods. By biochemical methods V. parahaemolyticus was detected in 55% of the samples, the highest being found in shellfish. The higher incidence of V. parahaemolyticus in shrimps can be correlated with the ability of this bacterium to utilize chitin, which is found abundant in shellfish.
Identification by biochemical method was initiated with the selection of sucrose followed by cellobiose non-fermenting colonies, and progressed with the other biochemical and microbiological tests as listed in the standard methods (FDA 1992). Earlier studies suggested that majority of the sucrose and cellobiose non-fermenting colonies could be V. vulnificus and V. campbelli other than V. parahaemolyticus (Elliot et al. 1995). The early elimination of vibrios other than these three vibrios by selecting sucrose/cellobiose non-fermenting colonies considerably reduced the time and labor involved in screening of V. parahaemolyticus isolates. It was reported that cellobiose fermentation can distinguish V. parahaemolyticus from other vibrios (Thampuran and Surendran 1998). In the present study all the isolates were subjected to cellobiose fermentation test, which could further narrow down the process of identification of presumable V. parahaemolyticus colonies. The sucrose and cellobiose non-fermenting colonies were selected and subjected to a battery of biochemical tests. Some colonies exhibited deviation from the standard methodology (Dileep et al. 2003) in giving few biochemical reactions. Those isolates which deviated in giving the reactions from the usual procedure are termed as atypical isolates, and identification of such isolates pose a major challenge for the microbiologists (Bharathi et al. 1987; Karunasagar et al. 1997). In the present study nine atypical isolates have been found, which deviated in the utilization of few sugars (arabinose, salicin, lactose, and inositol) and amino acids (lysine, arginine, and ornithine). Five of these atypical isolates were confirmed to be V. parahaemolyticus by targeting species-specific toxR (368 bp) gene.
All the biochemically confirmed isolates (55% of total isolates) were found to possess toxR (368 bp) gene. Moreover an additional 24% of the samples enriched in APW broth were confirmed to be positive for the presence of V. parahaemolyticus by this species-specific toxR gene. Thus the toxR targeted PCR method was found to be sensitive and rapid in the detection of V. parahaemolyticus from environmental samples as well as atypical colonies. The superiority of the toxR targeted PCR over the biochemical method was supported by earlier studies (Dileep et al. 2003).
In general, the phenotypic character such as urease activity corresponds to the presence of trh, a virulent gene as reported earlier (Shirai et al. 1990). In the present study two of the isolates were found to exhibit urease activity, and 50% of these urease positive isolates amplified trh (500 bp) gene. However, the remaining urease positive isolates did not amplify the trh gene. These results are in contrast with the studies of Okuda et al. (1997a, b) who reported the presence of trh gene in urease negative isolates. This observation led us to conclude that urease test alone may not be suitable for correlating the presence of trh gene in V. parahaemolyticus, and the results must be validated by PCR targeting trh gene.
The pathogenic V. parahaemolyticus isolates were found to exhibit β-haemolysis on blood agar, termed as Kanagawa positives (KP+), and such isolates were found to amplify the virulent gene tdh (269 bp). In the present study, 10% of the isolates were found to exhibit weak haemolysis (Kanagawa negative) and none of them were found to exhibit positive amplification for the tdh gene. The direct correlation between the presence of tdh gene and Kanagawa reaction led us to conclude that both tests are mandatory for the confirmation of pathogenicity of V. parahaemolyticus. The weak haemolysis points towards the presence of other virulence factors other than tdh in V. parahaemolyticus. The capability of the strains to produce few extra cellular enzymes may also lead to the weak haemolysis (Lee et al. 2002). There are reports of Kanagawa phenomenon being correlated with the positive detection of urease by urea hydrolysis (Kaysner et al. 1994; Iida et al. 1998). However, in the present study none of the weak haemolytic strains were found to exhibit urease activity.
Another gene orf8 was found to be associated with the virulence, was reported to be an useful genetic marker for the detection of pandemic V. parahaemolyticus strains. A unique serovar of V. parahaemolyticus O3:K6 possessing a unique open reading frame, orf8 was reported to be responsible for several outbreaks (Matsumoto et al. 2000). Although in the present study all the isolates were found to possess toxR gene, none of them were found to amplify the orf8 gene thus indicating the meager occurrence of pandemic strains in seafoods marketed from this area.
In conclusion, the present study emphasizes the importance of molecular method like PCR using enrichment broth lysates for identifying both pathogenic and non-pathogenic isolates exhibiting atypical biochemical reactions. This study provides the first direct evidence for the presence of pathogenic trh positive V. parahaemolyticus strains in the marketed seafood samples from the areas in and around Cochin. However, the lower incidence of trh positive pathogenic isolates and the complete absence of tdh and orf8 strains indicate that the seafoods marketed from Cochin are safe for consumption. In summary, this investigation thrusts upon the need for enhancing seafood-borne disease control strategies, focusing the attention on pathogenic V. parahaemolyticus strains, which are often linked with acute infections, associated with seafood ingestion.
References
Andrews WH, Hammack TS (2001) Salmonella. In: Bacteriological Analytical Manual Online, Chapter 5, 9th edn. US FDA Center for Food Safety and Applied Nutrition. Dedicated website at: http:// vm.cfsan.fda.gov/ebam/bam-toc.html
Ausubel FM, Brent R, Kingston RE, Moore DD, Sideman J, Smith J, Struhl K (1987) Current protocols in molecular biology. Wiley, New York
Bej AK, Patterson DP, Brasher CW, Vickery MCL, Jones DD, Kaysner CA (1999) Detection of total and hemolysin-producing Vibrio parahaemolyticus in shellfish using multiplex PCR amplification of tl, tdh and trh. J Microbiol Methods 36:215–225
Bharathi LPA, Nair S, Chandramohan D (1987) Occurrence and distribution of Vibrio parahaemolyticus and related organisms in the Arabian Sea. Mahasagar 20:43–51
Chowdhury NR, Chakraborty S, Ramamurthy T, Nishibuchi M, Yamasaki S, Takeda Y, Nair GB (2000) Molecular evidence of clonal Vibrio parahaemolyticus pandemic strains. Emerg Infect Dis 6:631–636
Dileep V, Kumar HS, Kumar Y, Nishibuchi M, Karunasagar I, Karunasagar I (2003) Application of polymerase chain reaction for detection of Vibrio parahaemolyticus associated with tropical seafoods and coastal environment. Lett Appl Microbiol 36:423–427
Elliot EL, Kaysner CA, Tamplin ML (1992) Media and reagents. In: USFDA bacteriological analytical manual, 7th edn. AOAC International, Arlington, Va, pp 508
Elliot EL, Kaysner CA, Jackson L, Tamplin ML (1995) V. cholerae, V. parahaemolyticus, V. vulnificus and other Vibrio spp. In: USFDA bacteriological analytical manual, 8th edn. AOAC International, Gaithersburg, MD, pp 901–927
Food, Drug Administration (1992) Bacteriological Analytical Manual, 2nd edn. Association of official analytical chemists, Washington DC
Fujino T, Okuno Y, Nakada D, Aoyama A, Fukai K, Mukai T, Ueho T (1953) On the bacteriological examination of shirasu food poisoning. Med J Osaka Univ 4:299–304
Hervio-Heath D, Colwell RR, Derrien A, Robert-Pillot A, Fournier JM, Pommepuy M (2002) Occurrence of pathogenic vibrios in coastal areas of France. J Appl Microbiol 92:1123–1135
Huss HH (1997) Control of indigenous pathogenic bacteria in seafood. Food Control 8:91–98
Iida T, Park KS, Suthienkul O, Kozawa J, Yamaichi Y, Yamamoto K, Honda T (1998) Close proximity of the tdh, trh and ure genes on the chromosome of Vibrio parahaemolyticus. Microbiology 144:2517–2523
Iida T, Hattori A, Tagomori K, Nasu H, Nair R, Honda T (2001) Filamentous phage associated with recent pandemic strains of Vibrio parahaemolyticus. Emer Infect Dis 7:477–478
Jacksic S, Suncica U, Petrak T, Bazulic D, Lada GK (2002) Occurrence of Vibrio spp. In: Sea fish, shrimps and bivalve mollusks harvested from Adriatic sea. Food Control 13:491–493
Joseph SW, Colwell RR, Kaper JB (1983) Vibrio parahaemolyticus and related halophilic vibrios. CRC Crit Rev Microbiol 10:77–123
Karunasagar I, Nayak BB, Karunasagar I (1997) Rapid detection of Vibrio parahaemolyticus from fish by polymerase chain reaction (PCR). In: Flegel T, MacRae IH (eds) Diseases in Asian aquaculture, 3rd edn. Asian Fisheries Society, Manila, pp 119–122
Kaysner CA, Abeyta C, Trost Jr PA, Wetherington JH, Jinneman KC, Hill WE, Wekell MM (1994) Urea hydrolysis can predict the potential pathogenicity of Vibrio parahaemolyticus strains isolated in the Pacific Northwest. Appl Environ Microbiol 60:3020–3022
Kim YB, Okuda J, Matsumoto C, Takahashi N, Hashimoto S, Nishibuchi M (1999) Identification of Vibrio parahaemolyticus strains at the species level by PCR targeted to the toxR gene. J Clin Microbiol 37:1173–1177
Kishishita M, Matsuoka N, Kumagai K, Yamasaki S, Takeda Y, Nishibuchi M (1992) Sequence variation in the thermostable direct hemolysin related hemolysin (trh) gene of Vibrio parahaemolyticus. Appl Environ Microbiol 58:2449
Lee CY, Cheng MF, Yu MS, Pan MJ (2002) Purification and characterization of a putative virulence factor, serine protease, from Vibrio parahaemolyticus. FEMS Microbiol Letts 209:31–37
Matsumoto C, Okuda J, Ishibashi M, Iwanaga M, Garg P, Ramamurthy T, Wong HC, De Paola A, Kim YB, Albert MJ, Nishibuchi M (2000) Pandemic spread of a O3:K6 clone of Vibrio parahaemolyticus and emergence of related strains evidenced by arbitrarily primed PCR and toxRS sequence analyses. J Clin Microbiol 38:578–585
Nasu H, Iida T, Sugahara Y, Yamaichi KS, Park K, Yokoyama K, Makino H, Shinagawa H, Honda T (2000) A filamentous phage associated with recent pandemic Vibrio parahaemolyticus O3:K6 strains. J Clin Microbiol 38:2156–2161
Okuda J, Ishibashi M, Abbott SL, Janda JM, Nishibuchi M (1997a) Analysis of the thermostable direct hemolysin (tdh) gene and the tdh-related hemolysin (trh) genes in urease-positive strains of Vibrio parahaemolyticus isolated on the West Coast of the United States. J Clin Microbiol 35:1965–1971
Okuda J, Ishibashi M, Hayakawa E, Nishino T, Takeda Y, Mukhopadhyay AK, Garg S, Bhattacharya SK, Nair GB, Nishibuchi M (1997b) Emergence of a unique O3:K6 clone of Vibrio parahaemolyticus in Calcutta, India, and isolation of strains from the same clonal group from Southeast Asian travelers arriving in Japan. J Clin Microbiol 35:3150–3155
Sanjeev S, Stephen J (1992) Antibiotic sensitivity of Kanagawa positive and Kanagawa-negative strains of Vibrio parahaemolyticus isolated from fishes marketed in Kochi. Fish Technol 29:162–165
Shirai H, Ito H, Hirayama T, Nakamoto Y, Nakabayashi N, Kumagai K, Takeda Y, Nishibuchi M (1990) Molecular evidence for association of the thermostable direct hemolysin (TDH) and TDH- related hemolysin of Vibrio parahaemolyticus with gastroenteritis. Infect Immun 58:3568–3573
Tada J, Ohashi T, Nishimura N, Shirasaki Y, Ozaki H, Fukushima S, Takano J, Nishibuchi M, Takeda Y (1992) Detection of thermostable direct hemolysin gene (tdh) and thermostable direct-related hemolysin gene (trh) of Vibrio parahaemolyticus by polymerase chain reaction. Mol Cell Prob 64:477–487
Thampuran N, Surendran PK (1998) Occurrence and distribution of Vibrio vulnificus in tropical fishes and shellfishes from Cochin (India). Lett Appl Microbiol 26:110–113
Wagatsuma S (1968) A medium for the test of the hemolytic activity of Vibrio parahaemolyticus. Media Circle 13:159–162
Waldor MK, Mekalanos JJ (1996) Lysogenic conversion by a filamentous phage encoding cholera toxin. Science 272:1910–1914
Wong HC, Chen MC, Liu SH, Liu DP (1999) Incidence of highly genetically diversified Vibrio parahaemolyticus in seafood imported from Asian countries. Int J Food Microbiol 52:181–188
Acknowledgements
The authors are grateful to Dr. K. Devadasan, Director, Central Institute of Fisheries Technology, Cochin and Prof. (Dr.) Mohan Joseph Modayil, Director, Central Marine Fisheries Research Institute for providing necessary facilities and encouragement for carrying out the study. The guidance and support rendered by Dr. Nirmala Thampuran, Head of the Mircrobiology Fermentation Biotechnology Division, Central Institute of Fisheries Technology, Cochin, to carry out this work are gratefully acknowledged.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Chakraborty, R.D., Surendran, P.K. Occurrence and distribution of virulent strains of Vibrio parahaemolyticus in seafoods marketed from Cochin (India). World J Microbiol Biotechnol 24, 1929–1935 (2008). https://doi.org/10.1007/s11274-008-9698-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11274-008-9698-2