Abstract
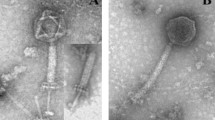
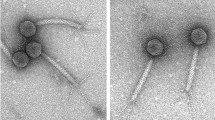
In the present study, two new Bacillus subtilis phages, BSTP4 and BSTP6, were isolated and studied further. Morphologically, BSTP4 and BSTP6 are podoviruses with complete genome of 19,145 (39.9% G + C content) and 19,367 bp (39.8% G + C content), respectively, which became among the smallest Bacillus phages. Three most prominent structural proteins were separated and identified as pre-neck appendage, major head, and head fiber proteins using LC–MS/MS. Both phages encode putative terminal proteins (TP) and contain short inverted terminal repeats (ITRs) which may be important for their replication. In addition, non-coding RNA (pRNA) and parS sites were identified which may be required for DNA packaging and their maintenance inside the host, respectively. Furthermore, the phage genome sequences show significant similarity to B. subtilis group species genome sequences. Finally, phylogenomic and phylogenetic analyses suggest that BSTP4 and BSTP6 may form a new species in the genus Salasvirus, subfamily Picovirinae of family Salasmaviridae. Considering the small numbers of ICTV-accepted B. subtilis phages and the importance of the host in the food industry and biotechnology, the current study helps to improve our understanding of the diversity of B. subtilis phages and shed light on the phage–host relationships.






Similar content being viewed by others
Data availability
The genomes of the phages are deposited in NCBI with GenBank accession number MW354668.1 for BSTP4 and MW354670.1 for BSTP6.
References
Tan YX, Mok WK, Lee J, Kim J, Chen WN (2019) Solid state fermentation of brewers’ spent grains for improved nutritional profile using Bacillus subtilis WX-17. Ferment 5(3):52
Chen JY, Yu YH (2021) Bacillus subtilis–fermented products ameliorate the growth performance and alter cecal microbiota community in broilers under lipopolysaccharide challenge. Poult Sci 100(2):875–886
Catalão MJ, Gil F, Moniz-Pereira J, São-José C, Pimentel M (2013) Diversity in bacterial lysis systems: bacteriophages show the way. FEMS Microbiol Rev 37(4):554–571
Chevallereau A, Pons BJ, van Houte S, Westra ER (2022) Interactions between bacterial and phage communities in natural environments. Nat Rev Microbiol 20(1):49–62
Obeng N, Pratama AA, van Elsas JD (2016) The significance of mutualistic phages for bacterial ecology and evolution. Trends Microbiol 24(6):440–449
O’Sullivan L, Bolton D, McAuliffe O, Coffey A (2022) The use of bacteriophages to control and detect pathogens in the dairy industry. Int J Dairy Technol. https://doi.org/10.1111/1471-0307.12641
Nagai T, Research FYF science and technology, 2009 U. Bacillus subtilis (natto) bacteriophages isolated in Japan. jstage.jst.go.jp [Internet]. 2009 [cited 2022 Oct 5]; Available from: https://www.jstage.jst.go.jp/article/fstr/15/3/15_3_293/_article/-char/ja/
Kubo Y, Sriyam S, Nakagawa R, Kimura KA (2018) Survey of phage contamination in natto-producing factories and development of phage-resistant Bacillus subtilis (natto) strains. Food Sci Technol Res 24(3):485–92
Bandara N, Jo J, Ryu S, Kim KP (2012) Bacteriophages BCP1-1 and BCP8-2 require divalent cations for efficient control of Bacillus cereus in fermented foods. Food Microbiol 31(1):9–16
Shin H, Bandara N, Shin E, Ryu S, Kim K, pyo (2011) Prevalence of Bacillus cereus bacteriophages in fermented foods and characterization of phage JBP901. Res Microbiol. 162(8):791–7
Ghosh K, Senevirathne A, Kang H, Hyun W, Viruses JK (2018) Complete nucleotide sequence analysis of a novel Bacillus subtilis-infecting bacteriophage BSP10 and its effect on poly-gamma-glutamic acid degradation. mdpi.com. 10(5):240
Zerbino DR, Birney E (2008) Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res 18(5):821–829
Delcher A, Harmon D, Kasif S, … OWN acids, 1999 U. Improved microbial gene identification with GLIMMER. academic.oup.com [Internet]. 1999 [cited 2022 Oct 5]; Available from: https://academic.oup.com/nar/article-abstract/27/23/4636/1064678
Hyatt D, Chen GL, LoCascio PF, Land ML, Larimer FW, Hauser LJ (2010) Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinform 8:11
Mcnair K, Zhou C, Dinsdale EA, Souza B, Edwards RA (2019) PHANOTATE: a novel approach to gene identification in phage genomes. Bioinformatics 35(22):4537–4542
Aziz RK, Bartels D, Best A, DeJongh M, Disz T, Edwards RA et al (2008) The RAST Server: rapid annotations using subsystems technology. BMC Genomics 8:9
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215(3):403–410
Eddy SR (2011) Accelerated profile HMM searches. PLoS Comput Biol. 7(10):1002195
Ecale Zhou CL, Malfatti S, Kimbrel J, Philipson C, McNair K, Hamilton T et al (2019) MultiPhATE: bioinformatics pipeline for functional annotation of phage isolates. Bioinformatics 35(21):4402–4404
Laslett D, Canback BARAGORN (2004) a program to detect tRNA genes and tmRNA genes in nucleotide sequences. Nucleic Acids Res 32(1):11–6
Perkins D, Pappin D, … DCE, 1999 U. Probability‐based protein identification by searching sequence databases using mass spectrometry data. Wiley Online Libr [Internet]. 1999 [cited 2022 Oct 5]; Available from: https://analyticalsciencejournals.onlinelibrary.wiley.com/doi/abs/https://doi.org/10.1002/(SICI)1522-2683(19991201)20:18%3C3551::AID-ELPS3551%3E3.0.CO;2-2
Lefkowitz EJ, Dempsey DM, Hendrickson RC, Orton RJ, Siddell SG, Smith DB. Virus taxonomy: the database of the International Committee on Taxonomy of Viruses (ICTV). Nucleic Acids Res [Internet]. 2018 Jan 1 [cited 2022 Oct 5];46(D1):D708–17. Available from: https://pubmed.ncbi.nlm.nih.gov/29040670/
Ågren J, Sundström A, Håfström T, Segerman B (2012) Gegenees: Fragmented alignment of multiple genomes for determining phylogenomic distances and genetic signatures unique for specified target groups. PLoS One. 7(6):e39107
Sullivan MJ, Petty NK, Beatson SA (2011) Easyfig: a genome comparison visualizer. Bioinformatics 27(7):1009–1010
Dereeper A, Guignon V, Blanc G, Audic S, Buffet S, Chevenet F et al (2008) Phylogenyfr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res 36:W465–W469
Thompson JD, Gibson TJ, Higgins DG (2003) Multiple sequence alignment using ClustalW and ClustalX. Curr Protoc Bioinforma. 1:2–3
Kumar S, Stecher G, Tamura K (2016) MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33(7):1870–4
Meijer WJJ, Horcajadas JA, Salas M (2001) φ29 Family of Phages. Microbiol Mol Biol Rev 65(2):261–287
Schilling T, Hoppert M, Daniel R, Hertel R (2018) Complete genome sequence of vB_BveP-Goe6 a virus infecting Bacillus velezensis FZB42. Genome Announc 6(8):e00008-18
Guo X, Zhang T, Jin M, Zeng R (2020) Characterization of Bacillus phage Gxv1, a novel lytic Salasvirus phage isolated from deep-sea seamount sediments. Mar Life Sci. https://doi.org/10.1007/s42995-020-00074-8
Pačes V, Viček Č, Urbánek P, Hostomský Z (1985) Nucleotide sequence of the major early region of Bacillus subtilis phage PZA, a close relative of φ29. Gene 38(1–3):45–56
Willms IM, Hertel R (2016) Phage vB_BsuP-Goe1: the smallest identified lytic phage of Bacillus subtilis. FEMS Microbiol Lett 363(19):fnw208
Bradley DE (1965) The isolation and morphology of some new bacteriophages specific for Bacillus and Acetobacter species. J Gen Microbiol 41(2):233–241
Taylor AL (2022) Bacteriophage-induced mutation in Escherichia Coli. Proc Natl Acad Sci USA 50(2):1043–51
Wang PW, Chu L, Guttman DS (2004) Complete sequence and evolutionary genomic analysis of the Pseudomonas aeruginosa transposable bacteriophage D3112. J Bacteriol 186(2):400–410
Wu H, Zhang Y, Jiang Y, Wu H, Sun W, Huang YP (2021) Characterization and genomic analysis of ɸSHP3, a new transposable bacteriophage infecting Stenotrophomonas Maltophilia. J Virol. 95(9):e00019-21
Meijer WJ, Castilla-Llorente V, Villar L, Murray H, Errington J, Salas M (2005) Molecular basis for the exploitation of sporeformation as survival mechanism by virulent phage ϕ29. EMBO J 24(20):3647–3657
Castilla-Llorente V, Meijer WJJ, Salas M (2009) Differential Spo0A-mediated effects on transcription and replication of the related Bacillus subtilis phages Nf and Φ29 explain their different behaviours in vivo. Nucleic Acids Res 37(15):4955–4964
Castilla-Llorente V, Salas M, Meijer WJJ (2022) kinC/D-mediated heterogeneous expression of spo0A during logarithmical growth in Bacillus subtilis is responsible for partial suppression of φ29 development. Wiley 68(6):1406–17. https://doi.org/10.1111/j.1365-2958.2008.06234.x
Meijer WJJ, Castilla-Llorente V, Villar L, Murray H, Errington J, Salas M (2005) Molecular basis for the exploitation of spore formation as survival mechanism by virulent phage φ29. EMBO J 24(20):3647–3657
Motlagh AM, Bhattacharjee AS, Coutinho FH, Dutilh BE, Casjens SR, Goel RK (2017) Insights of phage-host interaction in hypersaline ecosystem through metagenomics analyses. Front Microbiol. 3:8–352
Turner D, Kropinski AM, Adriaenssens EM. A Roadmap for Genome-Based Phage Taxonomy. Viruses [Internet]. 2021 Mar 1 [cited 2023 Jan 6];13(3). Available from: https://pubmed.ncbi.nlm.nih.gov/33803862/
Walker PJ, Siddell SG, Lefkowitz EJ, Mushegian AR, Adriaenssens EM, Dempsey DM et al (2020) Changes to virus taxonomy and the Statutes ratified by the International Committee on Taxonomy of Viruses (2020). Arch Virol 165(11):2737–2748
Yoshikawa H, Ito J (1981) Terminal proteins and short inverted terminal repeats of the small Bacillus bacteriophage genomes. Proc Natl Acad Sci USA 78(4):2596–600
Bjornsti MA, Reilly BE, Anderson DL (1983) Morphogenesis of bacteriophage phi 29 of Bacillus subtilis: oriented and quantized in vitro packaging of DNA protein gp3. J Virol 45(1):383–396
Adriaenssens EM, Rodney Brister J (2017) How to name and classify your phage: an informal guide. Viruses 9(4):70
Ripp S, Miller RV (1997) The role of pseudolysogeny in bacteriophage-host interactions in a natural freshwater environment. Microbiology 143(6):2065–2070
Siringan P, Connerton PL, Cummings NJ, Connerton IF (2014) Alternative bacteriophage life cycles: the carrier state of campylobacter jejuni. Open Biol. 4(3):130200
Łoś M, Węgrzyn G. Chapter 9 – Pseudolysogeny [Internet]. Vol. 82, Advances in Virus Research. 2012. p. 339–49. Available from: http://www.sciencedirect.com.ezproxy.weizmann.ac.il/science/article/pii/B9780123946218000194
Casjens S (2003) Prophages and bacterial genomics: what have we learned so far? Mol Microbiol 49(2):277–300
Howard-Varona C, Hargreaves KR, Abedon ST, Sullivan MB (2017) Lysogeny in nature: mechanisms impact and ecology of temperate phages. ISME J 11(7):1511
Grau RR, De Oña P, Kunert M, Leñini C, Gallegos-Monterrosa R, Mhatre E et al (2015) A duo of potassium-responsive histidine kinases govern the multicellular destiny of Bacillus subtilis. MBio. 6(4):00581–15
Acknowledgements
We are grateful to Jeonbuk National University (JBNU) for resource support as well as the National Research Foundation of Korea (NRF), Korea Government (MSIT), and Global Korea Scholarship Program [GKS] for financial support.
Funding
This work was supported by the National Research Foundation of Korea (NRF) grant funded by the Korea Government (MSIT) [grant number: 2021R1A2C2008022].
Author information
Authors and Affiliations
Contributions
KPK and HBA conceived and designed the experiments. KPK supervised the experiment and critically reviewed the manuscript for its enrichment. HBA did the experiment, analysis, and wrote the draft of the manuscript. Both authors have read and agreed to the published version of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Edited by Andrew Millard.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Abraha, H.B., Kim, KP. Complete genome sequence analysis, morphology and structural protein identification of two Bacillus subtilis phages, BSTP4 and BSTP6, which may form a new species in the genus Salasvirus. Virus Genes 59, 624–634 (2023). https://doi.org/10.1007/s11262-023-01998-w
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11262-023-01998-w