Abstract
The Jumonji C-domain containing protein 6 (JMJD6) has had a convoluted history, and recent reports indicating a multifactorial role in foot-and-mouth disease virus (FMDV) infection have further complicated the functionality of this protein. It was first identified as the phosphatidylserine receptor on the cell surface responsible for recognizing phosphatidylserine on the surface of apoptotic cells resulting in their engulfment by phagocytic cells. Subsequent study revealed a nuclear subcellular localization, where JMJD6 participated in lysine hydroxylation and arginine demethylation of histone proteins and other non-histone proteins. Interestingly, to date, JMDJ6 remains the only known arginine demethylase with a growing list of known substrate molecules. These conflicting associations rendered the subcellular localization of JMJD6 to be quite nebulous. Further muddying this area, two different groups illustrated that JMJD6 could be induced to redistribute from the cell surface to the nucleus of a cell. More recently, JMJD6 was demonstrated to be a host factor contributing to the FMDV life cycle, where it was not only exploited for its arginine demethylase activity, but also served as an alternative virus receptor. This review attempts to coalesce these divergent roles for a single protein into one cohesive account. Given the diverse functionalities already characterized for JMJD6, it is likely to continue to be a confounding protein resulting in much contention going into the near future.



Similar content being viewed by others
References
H. Hou, H. Yu, Structural insights into histone lysine demethylation. Curr. Opin. Struct. Biol. 20, 739–748 (2010)
T.B. Nicholson, T. Chen, LSD1 demethylates histone and non-histone proteins. Epigenetics 4, 129–132 (2009)
S.M. Hensen, G.J. Pruijn, Methods for the detection of peptidylarginine deiminase (PAD) activity and protein citrullination. Mol. Cell. Proteomics 13, 388–396 (2014)
B. Chang, Y. Chen, Y. Zhao, R.K. Bruick, JMJD6 is a histone arginine demethylase. Science 318, 444–447 (2007)
J. Motoyama, T. Takeuchi, The mouse embryogenesis of jumonji mutant obtained by gene-trap method. Tanpakushitsu Kakusan Koso 40, 2152–2161 (1995)
T. Takeuchi, Y. Yamazaki, Y. Katoh-Fukui, R. Tsuchiya, S. Kondo, J. Motoyama, T. Higashinakagawa, Gene trap capture of a novel mouse gene, jumonji, required for neural tube formation. Genes Dev. 9, 1211–1222 (1995)
M. Cikala, O. Alexandrova, C.N. David, M. Proschel, B. Stiening, P. Cramer, A. Bottger, The phosphatidylserine receptor from Hydra is a nuclear protein with potential Fe(II) dependent oxygenase activity. BMC Cell Biol. 5, 26 (2004)
A. Wolf, M. Mantri, A. Heim, U. Muller, E. Fichter, M.M. Mackeen, L. Schermelleh, G. Dadie, H. Leonhardt, C. Venien-Bryan, B.M. Kessler, C.J. Schofield, A. Bottger, The polyserine domain of the lysyl-5 hydroxylase Jmjd6 mediates subnuclear localization. Biochem. J. 453, 357–370 (2013)
P. Cui, B. Qin, N. Liu, G. Pan, D. Pei, Nuclear localization of the phosphatidylserine receptor protein via multiple nuclear localization signals. Exp. Cell Res. 293, 154–163 (2004)
H. Yang, Y.Z. Chen, Y. Zhang, X. Wang, X. Zhao, J.I. Godfroy 3rd, Q. Liang, M. Zhang, T. Zhang, Q. Yuan, M. Ann Royal, M. Driscoll, N.S. Xia, H. Yin, D. Xue, A lysine-rich motif in the phosphatidylserine receptor PSR-1 mediates recognition and removal of apoptotic cells. Nat. Commun. 6, 5717 (2015)
P. Hahn, J. Bose, S. Edler, A. Lengeling, Genomic structure and expression of Jmjd6 and evolutionary analysis in the context of related JmjC domain containing proteins. BMC Genom. 9, 293 (2008)
V.A. Fadok, D.L. Bratton, D.M. Rose, A. Pearson, R.A. Ezekewitz, P.M. Henson, A receptor for phosphatidylserine-specific clearance of apoptotic cells. Nature 405, 85–90 (2000)
V.A. Fadok, A. de Cathelineau, D.L. Daleke, P.M. Henson, D.L. Bratton, Loss of phospholipid asymmetry and surface exposure of phosphatidylserine is required for phagocytosis of apoptotic cells by macrophages and fibroblasts. J. Biol. Chem. 276, 1071–1077 (2001)
V.A. Fadok, D. Xue, P. Henson, If phosphatidylserine is the death knell, a new phosphatidylserine-specific receptor is the bellringer. Cell Death Differ. 8, 582–587 (2001)
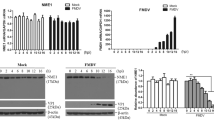
P. Lawrence, J. Pacheco, C. Stenfeldt, J. Arzt, D.K. Rai, E. Rieder, Pathogenesis and micro-anatomic characterization of a cell-adapted mutant foot-and-mouth disease virus in cattle: impact of the Jumonji C-domain containing protein 6 (JMJD6) and route of inoculation. Virology 492, 108–117 (2016)
P. Lawrence, D. Rai, J.S. Conderino, S. Uddowla, E. Rieder, Role of Jumonji C-domain containing protein 6 (JMJD6) in infectivity of foot-and-mouth disease virus. Virology 492, 38–52 (2016)
P. Lawrence, J.S. Conderino, E. Rieder, Redistribution of demethylated RNA helicase A during foot-and-mouth disease virus infection: role of Jumonji C-domain containing protein 6 in RHA demethylation. Virology 452–453, 1–11 (2014)
A. Yamaji-Hasegawa, M. Tsujimoto, Asymmetric distribution of phospholipids in biomembranes. Biol. Pharm. Bull. 29, 1547–1553 (2006)
V.A. Fadok, D.L. Bratton, S.C. Frasch, M.L. Warner, P.M. Henson, The role of phosphatidylserine in recognition of apoptotic cells by phagocytes. Cell Death Differ. 5, 551–562 (1998)
P.R. Hoffmann, A.M. deCathelineau, C.A. Ogden, Y. Leverrier, D.L. Bratton, D.L. Daleke, A.J. Ridley, V.A. Fadok, P.M. Henson, Phosphatidylserine (PS) induces PS receptor-mediated macropinocytosis and promotes clearance of apoptotic cells. J. Cell Biol. 155, 649–659 (2001)
X. Wang, Y.C. Wu, V.A. Fadok, M.C. Lee, K. Gengyo-Ando, L.C. Cheng, D. Ledwich, P.K. Hsu, J.Y. Chen, B.K. Chou, P. Henson, S. Mitani, D. Xue, Cell corpse engulfment mediated by C. elegans phosphatidylserine receptor through CED-5 and CED-12. Science 302, 1563–1566 (2003)
T.Y. Hsu, Y.C. Wu, Engulfment of apoptotic cells in C. elegans is mediated by integrin alpha/SRC signaling. Curr. Biol. 20, 477–486 (2010)
X. Wang, C. Yang, Programmed cell death and clearance of cell corpses in Caenorhabditis elegans. Cell. Mol. Life Sci. 73, 2221–2236 (2016)
A.C. Tosello-Trampont, J.M. Kinchen, E. Brugnera, L.B. Haney, M.O. Hengartner, K.S. Ravichandran, Identification of two signaling submodules within the CrkII/ELMO/Dock180 pathway regulating engulfment of apoptotic cells. Cell Death Differ. 14, 963–972 (2007)
J.R. Hong, G.H. Lin, C.J. Lin, W.P. Wang, C.C. Lee, T.L. Lin, J.L. Wu, Phosphatidylserine receptor is required for the engulfment of dead apoptotic cells and for normal embryonic development in zebrafish. Development 131, 5417–5427 (2004)
H.C. Kung, O. Evensen, J.R. Hong, C.Y. Kuo, C.H. Tso, F.H. Ngou, M.W. Lu, J.L. Wu, Interferon regulatory factor-1 (IRF-1) is involved in the induction of phosphatidylserine receptor (PSR) in response to dsRNA virus infection and contributes to apoptotic cell clearance in CHSE-214 cell. Int. J. Mol. Sci. 15, 19281–19306 (2014)
P.M. Henson, D.L. Bratton, V.A. Fadok, The phosphatidylserine receptor: a crucial molecular switch? Nat. Rev. Mol. Cell Biol. 2, 627–633 (2001)
P.R. Hoffmann, J.A. Kench, A. Vondracek, E. Kruk, D.L. Daleke, M. Jordan, P. Marrack, P.M. Henson, V.A. Fadok, Interaction between phosphatidylserine and the phosphatidylserine receptor inhibits immune responses in vivo. J. Immunol. 174, 1393–1404 (2005)
S. Akakura, S. Singh, M. Spataro, R. Akakura, J.I. Kim, M.L. Albert, R.B. Birge, The opsonin MFG-E8 is a ligand for the alphavbeta5 integrin and triggers DOCK180-dependent Rac1 activation for the phagocytosis of apoptotic cells. Exp. Cell Res. 292, 403–416 (2004)
R. Hanayama, M. Tanaka, K. Miwa, A. Shinohara, A. Iwamatsu, S. Nagata, Identification of a factor that links apoptotic cells to phagocytes. Nature 417, 182–187 (2002)
V. D’Mello, R.B. Birge, Apoptosis: conserved roles for integrins in clearance. Curr. Biol. 20, R324–R327 (2010)
M.L. Albert, J.I. Kim, R.B. Birge, Alphavbeta5 integrin recruits the CrkII-Dock180-rac1 complex for phagocytosis of apoptotic cells. Nat. Cell Biol. 2, 899–905 (2000)
J. Savill, I. Dransfield, N. Hogg, C. Haslett, Vitronectin receptor-mediated phagocytosis of cells undergoing apoptosis. Nature 343, 170–173 (1990)
Y. Ishimoto, K. Ohashi, K. Mizuno, T. Nakano, Promotion of the uptake of PS liposomes and apoptotic cells by a product of growth arrest-specific gene, gas6. J. Biochem. 127, 411–417 (2000)
T. Hisatomi, T. Sakamoto, K.H. Sonoda, C. Tsutsumi, H. Qiao, H. Enaida, I. Yamanaka, T. Kubota, T. Ishibashi, S. Kura, S.A. Susin, G. Kroemer, Clearance of apoptotic photoreceptors: elimination of apoptotic debris into the subretinal space and macrophage-mediated phagocytosis via phosphatidylserine receptor and integrin alphavbeta3. Am. J. Pathol. 162, 1869–1879 (2003)
R.J. Krieser, F.E. Moore, D. Dresnek, B.J. Pellock, R. Patel, A. Huang, C. Brachmann, K. White, The Drosophila homolog of the putative phosphatidylserine receptor functions to inhibit apoptosis. Development 134, 2407–2414 (2007)
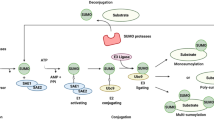
A. Bottger, M.S. Islam, R. Chowdhury, C.J. Schofield, A. Wolf, The oxygenase Jmjd6–a case study in conflicting assignments. Biochem. J. 468, 191–202 (2015)
G. Han, J. Li, Y. Wang, X. Li, H. Mao, Y. Liu, C.D. Chen, The hydroxylation activity of Jmjd6 is required for its homo-oligomerization. J. Cell. Biochem. 113, 1663–1670 (2012)
K.K. Penberthy, K.S. Ravichandran, Apoptotic cell recognition receptors and scavenger receptors. Immunol. Rev. 269, 44–59 (2016)
G.J. Freeman, J.M. Casasnovas, D.T. Umetsu, R.H. DeKruyff, TIM genes: a family of cell surface phosphatidylserine receptors that regulate innate and adaptive immunity. Immunol. Rev. 235, 172–189 (2010)
L. Meertens, X. Carnec, M.P. Lecoin, R. Ramdasi, F. Guivel-Benhassine, E. Lew, G. Lemke, O. Schwartz, A. Amara, The TIM and TAM families of phosphatidylserine receptors mediate dengue virus entry. Cell Host Microbe 12, 544–557 (2012)
M. Miyanishi, K. Tada, M. Koike, Y. Uchiyama, T. Kitamura, S. Nagata, Identification of Tim4 as a phosphatidylserine receptor. Nature 450, 435–439 (2007)
S. Moller-Tank, W. Maury, Phosphatidylserine receptors: enhancers of enveloped virus entry and infection. Virology 468–470, 565–580 (2014)
K. Morizono, I.S. Chen, Role of phosphatidylserine receptors in enveloped virus infection. J. Virol. 88, 4275–4290 (2014)
P. Hahn, I. Wegener, A. Burrells, J. Bose, A. Wolf, C. Erck, D. Butler, C.J. Schofield, A. Bottger, A. Lengeling, Analysis of Jmjd6 cellular localization and testing for its involvement in histone demethylation. PLoS One 5, e13769 (2010)
C.J. Webby, A. Wolf, N. Gromak, M. Dreger, H. Kramer, B. Kessler, M.L. Nielsen, C. Schmitz, D.S. Butler, J.R. Yates 3rd, C.M. Delahunty, P. Hahn, A. Lengeling, M. Mann, N.J. Proudfoot, C.J. Schofield, A. Bottger, Jmjd6 catalyses lysyl-hydroxylation of U2AF65, a protein associated with RNA splicing. Science 325, 90–93 (2009)
W.W. Gao, R.Q. Xiao, B.L. Peng, H.T. Xu, H.F. Shen, M.F. Huang, T.T. Shi, J. Yi, W.J. Zhang, X.N. Wu, X. Gao, X.Z. Lin, P.C. Dorrestein, M.G. Rosenfeld, W. Liu, Arginine methylation of HSP70 regulates retinoid acid-mediated RARbeta2 gene activation. Proc. Natl. Acad. Sci. USA 112, E3327–E3336 (2015)
C. Poulard, J. Rambaud, N. Hussein, L. Corbo, M. Le Romancer, JMJD6 regulates ERalpha methylation on arginine. PLoS One 9, e87982 (2014)
I. Tikhanovich, S. Kuravi, A. Artigues, M.T. Villar, K. Dorko, A. Nawabi, B. Roberts, S.A. Weinman, Dynamic arginine methylation of tumor necrosis factor (TNF) receptor-associated factor 6 regulates Toll-like receptor signaling. J. Biol. Chem. 290, 22236–22249 (2015)
T.F. Wu, Y.L. Yao, I.L. Lai, C.C. Lai, P.L. Lin, W.M. Yang, Loading of PAX3 to mitotic chromosomes is mediated by arginine methylation and associated with waardenburg syndrome. J. Biol. Chem. 290, 20556–20564 (2015)
F.V. Fuller-Pace, DExD/H box RNA helicases: multifunctional proteins with important roles in transcriptional regulation. Nucl. Acids Res. 34, 4206–4215 (2006)
F. Wang, L. He, P. Huangyang, J. Liang, W. Si, R. Yan, X. Han, S. Liu, B. Gui, W. Li, D. Miao, C. Jing, Z. Liu, F. Pei, L. Sun, Y. Shang, JMJD6 promotes colon carcinogenesis through negative regulation of p53 by hydroxylation. PLoS Biol. 12, e1001819 (2014)
J. Zhang, S.S. Ni, W.L. Zhao, X.C. Dong, J.L. Wang, High expression of JMJD6 predicts unfavorable survival in lung adenocarcinoma. Tumour Biol. 34, 2397–2401 (2013)
C. Poulard, J. Rambaud, E. Lavergne, J. Jacquemetton, J.M. Renoir, O. Tredan, S. Chabaud, I. Treilleux, L. Corbo, M. Le Romancer, Role of JMJD6 in breast tumourigenesis. PLoS One 10, e0126181 (2015)
Y.F. Lee, L.D. Miller, X.B. Chan, M.A. Black, B. Pang, C.W. Ong, M. Salto-Tellez, E.T. Liu, K.V. Desai, JMJD6 is a driver of cellular proliferation and motility and a marker of poor prognosis in breast cancer. Breast Cancer Res. 14, R85 (2012)
O. Aprelikova, K. Chen, L.H. El Touny, C. Brignatz-Guittard, J. Han, T. Qiu, H.H. Yang, M.P. Lee, M. Zhu, J.E. Green, The epigenetic modifier JMJD6 is amplified in mammary tumors and cooperates with c-Myc to enhance cellular transformation, tumor progression, and metastasis. Clin. Epigenet. 8, 38 (2016)
C.R. Lee, S.H. Lee, N.K. Rigas, R.H. Kim, M.K. Kang, N.H. Park, K.H. Shin, Elevated expression of JMJD6 is associated with oral carcinogenesis and maintains cancer stemness properties. Carcinogenesis 37, 119–128 (2016)
J. Wan, W. Xu, J. Zhan, J. Ma, X. Li, Y. Xie, J. Wang, W.G. Zhu, J. Luo, H. Zhang, PCAF-mediated acetylation of transcriptional factor HOXB9 suppresses lung adenocarcinoma progression by targeting oncogenic protein JMJD6. Nucl. Acids Res. (2016). doi:10.1093/nar/gkw808
C. Poulard, L. Corbo, M. Le Romancer, Protein arginine methylation/demethylation and cancer. Oncotarget (2016). doi:10.18632/oncotarget.11376
L.J. Walport, R.J. Hopkinson, R. Chowdhury, R. Schiller, W. Ge, A. Kawamura, C.J. Schofield, Arginine demethylation is catalysed by a subset of JmjC histone lysine demethylases. Nat. Commun. 7, 11974 (2016)
A. Heim, C. Grimm, U. Muller, S. Haussler, M.M. Mackeen, J. Merl, S.M. Hauck, B.M. Kessler, C.J. Schofield, A. Wolf, A. Bottger, Jumonji domain containing protein 6 (Jmjd6) modulates splicing and specifically interacts with arginine-serine-rich (RS) domains of SR- and SR-like proteins. Nucl. Acids Res. 42, 7833–7850 (2014)
J. Barman-Aksozen, C. Beguin, A.M. Dogar, X. Schneider-Yin, E.I. Minder, Iron availability modulates aberrant splicing of ferrochelatase through the iron- and 2-oxoglutarate dependent dioxygenase Jmjd6 and U2AF(65.). Blood Cells Mol. Dis. 51, 151–161 (2013)
M. Mantri, T. Krojer, E.A. Bagg, C.J. Webby, D.S. Butler, G. Kochan, K.L. Kavanagh, U. Oppermann, M.A. McDonough, C.J. Schofield, Crystal structure of the 2-oxoglutarate- and Fe(II)-dependent lysyl hydroxylase JMJD6. J. Mol. Biol. 401, 211–222 (2010)
X. Hong, J. Zang, J. White, C. Wang, C.H. Pan, R. Zhao, R.C. Murphy, S. Dai, P. Henson, J.W. Kappler, J. Hagman, G. Zhang, Interaction of JMJD6 with single-stranded RNA. Proc. Natl. Acad. Sci. USA 107, 14568–14572 (2010)
M. Unoki, A. Masuda, N. Dohmae, K. Arita, M. Yoshimatsu, Y. Iwai, Y. Fukui, K. Ueda, R. Hamamoto, M. Shirakawa, H. Sasaki, Y. Nakamura, Lysyl 5-hydroxylation, a novel histone modification, by Jumonji domain containing 6 (JMJD6). J. Biol. Chem. 288, 6053–6062 (2013)
N. Tibrewal, T. Liu, H. Li, R.B. Birge, Characterization of the biochemical and biophysical properties of the phosphatidylserine receptor (PS-R) gene product. Mol. Cell. Biochem. 304, 119–125 (2007)
L. Zakharova, S. Dadsetan, A.F. Fomina, Endogenous Jmjd6 gene product is expressed at the cell surface and regulates phagocytosis in immature monocyte-like activated THP-1 cells. J. Cell. Physiol. 221, 84–91 (2009)
M.J. Grubman, B. Baxt, Foot-and-mouth disease. Clin. Microbiol. Rev. 17, 465–493 (2004)
P.W. Mason, M.J. Grubman, B. Baxt, Molecular basis of pathogenesis of FMDV. Virus Res. 91, 9–32 (2003)
M. Garcia-Briones, M.F. Rosas, M. Gonzalez-Magaldi, M.A. Martin-Acebes, F. Sobrino, R. Armas-Portela, Differential distribution of non-structural proteins of foot-and-mouth disease virus in BHK-21 cells. Virology 349, 409–421 (2006)
P. Lawrence, E.A. Schafer, E. Rieder, The nuclear protein Sam68 is cleaved by the FMDV 3C protease redistributing Sam68 to the cytoplasm during FMDV infection of host cells. Virology 425, 40–52 (2012)
S. Neff, D. Sa-Carvalho, E. Rieder, P.W. Mason, S.D. Blystone, E.J. Brown, B. Baxt, Foot-and-mouth disease virus virulent for cattle utilizes the integrin alpha(v)beta3 as its receptor. J. Virol. 72, 3587–3594 (1998)
T. Jackson, S. Clark, S. Berryman, A. Burman, S. Cambier, D. Mu, S. Nishimura, A.M. King, Integrin alphavbeta8 functions as a receptor for foot-and-mouth disease virus: role of the beta-chain cytodomain in integrin-mediated infection. J. Virol. 78, 4533–4540 (2004)
T. Jackson, A.P. Mould, D. Sheppard, A.M. King, Integrin alphavbeta1 is a receptor for foot-and-mouth disease virus. J. Virol. 76, 935–941 (2002)
T. Jackson, D. Sheppard, M. Denyer, W. Blakemore, A.M. King, The epithelial integrin alphavbeta6 is a receptor for foot-and-mouth disease virus. J. Virol. 74, 4949–4956 (2000)
H. Duque, B. Baxt, Foot-and-mouth disease virus receptors: comparison of bovine alpha(V) integrin utilization by type A and O viruses. J. Virol. 77, 2500–2511 (2003)
H. Duque, M. LaRocco, W.T. Golde, B. Baxt, Interactions of foot-and-mouth disease virus with soluble bovine alphaVbeta3 and alphaVbeta6 integrins. J. Virol. 78, 9773–9781 (2004)
V. O’Donnell, M. LaRocco, H. Duque, B. Baxt, Analysis of foot-and-mouth disease virus internalization events in cultured cells. J. Virol. 79, 8506–8518 (2005)
S. Berryman, S. Clark, P. Monaghan, T. Jackson, Early events in integrin alphavbeta6-mediated cell entry of foot-and-mouth disease virus. J. Virol. 79, 8519–8534 (2005)
E. Baranowski, C.M. Ruiz-Jarabo, N. Sevilla, D. Andreu, E. Beck, E. Domingo, Cell recognition by foot-and-mouth disease virus that lacks the RGD integrin-binding motif: flexibility in aphthovirus receptor usage. J. Virol. 74, 1641–1647 (2000)
D. Sa-Carvalho, E. Rieder, B. Baxt, R. Rodarte, A. Tanuri, P.W. Mason, Tissue culture adaptation of foot-and-mouth disease virus selects viruses that bind to heparin and are attenuated in cattle. J. Virol. 71, 5115–5123 (1997)
V. O’Donnell, M. Larocco, B. Baxt, Heparan sulfate-binding foot-and-mouth disease virus enters cells via caveola-mediated endocytosis. J. Virol. 82, 9075–9085 (2008)
M.V. Borca, J.M. Pacheco, L.G. Holinka, C. Carrillo, E. Hartwig, D. Garriga, E. Kramer, L. Rodriguez, M.E. Piccone, Role of arginine-56 within the structural protein VP3 of foot-and-mouth disease virus (FMDV) O1 Campos in virus virulence. Virology 422, 37–45 (2012)
F.F. Maree, B. Blignaut, L. Aschenbrenner, T. Burrage, E. Rieder, Analysis of SAT1 type foot-and-mouth disease virus capsid proteins: influence of receptor usage on the properties of virus particles. Virus Res. 155, 462–472 (2011)
S. Berryman, S. Clark, N.K. Kakker, R. Silk, J. Seago, J. Wadsworth, K. Chamberlain, N.J. Knowles, T. Jackson, Positively charged residues at the five-fold symmetry axis of cell culture-adapted foot-and-mouth disease virus permit novel receptor interactions. J. Virol. 87, 8735–8744 (2013)
E.E. Fry, S.M. Lea, T. Jackson, J.W. Newman, F.M. Ellard, W.E. Blakemore, R. Abu-Ghazaleh, A. Samuel, A.M. King, D.I. Stuart, The structure and function of a foot-and-mouth disease virus-oligosaccharide receptor complex. EMBO J. 18, 543–554 (1999)
F.F. Maree, B. Blignaut, T.A. de Beer, N. Visser, E.A. Rieder, Mapping of amino acid residues responsible for adhesion of cell culture-adapted foot-and-mouth disease SAT type viruses. Virus Res. 153, 82–91 (2010)
T.S. McKenna, J. Lubroth, E. Rieder, B. Baxt, P.W. Mason, Receptor binding site-deleted foot-and-mouth disease (FMD) virus protects cattle from FMD. J. Virol. 69, 5787–5790 (1995)
Q. Zhao, J.M. Pacheco, P.W. Mason, Evaluation of genetically engineered derivatives of a Chinese strain of foot-and-mouth disease virus reveals a novel cell-binding site which functions in cell culture and in animals. J. Virol. 77, 3269–3280 (2003)
P. Lawrence, M. LaRocco, B. Baxt, E. Rieder, Examination of soluble integrin resistant mutants of foot-and-mouth disease virus. Virol. J. 10, 2 (2013)
M.E. Piccone, S. Sira, M. Zellner, M.J. Grubman, Expression in Escherichia coli and purification of biologically active L proteinase of foot-and-mouth disease virus. Virus Res. 35, 263–275 (1995)
J.D. Esko, K.S. Rostand, J.L. Weinke, Tumor formation dependent on proteoglycan biosynthesis. Science 241, 1092–1096 (1988)
M.E. Piccone, J.M. Pacheco, S.J. Pauszek, E. Kramer, E. Rieder, M.V. Borca, L.L. Rodriguez, The region between the two polyprotein initiation codons of foot-and-mouth disease virus is critical for virulence in cattle. Virology 396, 152–159 (2010)
A. Amara, J. Mercer, Viral apoptotic mimicry. Nat. Rev. Microbiol. 13, 461–469 (2015)
Y.H. Chen, W. Du, M.C. Hagemeijer, P.M. Takvorian, C. Pau, A. Cali, C.A. Brantner, E.S. Stempinski, P.S. Connelly, H.C. Ma, P. Jiang, E. Wimmer, G. Altan-Bonnet, N. Altan-Bonnet, Phosphatidylserine vesicles enable efficient en bloc transmission of enteroviruses. Cell 160, 619–630 (2015)
Z. Feng, Y. Li, K.L. McKnight, L. Hensley, R.E. Lanford, C.M. Walker, S.M. Lemon, Human pDCs preferentially sense enveloped hepatitis A virions. J. Clin. Investig. 125, 169–176 (2015)
M.F. Barber, N.C. Elde, Evolutionary biology: mimicry all the way down. Nature 501, 38–39 (2013)
V. O’Donnell, J.M. Pacheco, M. LaRocco, T. Burrage, W. Jackson, L.L. Rodriguez, M.V. Borca, B. Baxt, Foot-and-mouth disease virus utilizes an autophagic pathway during viral replication. Virology 410, 142–150 (2011)
Acknowledgements
The authors wish to thank Isaiah Negron and Joseph Gutkoska for their critical review of this manuscript with suggested modifications as well as for fruitful discussions.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
Dr. Paul Lawrence declares that he has no conflicts of interest. Dr. Elizabeth Rieder declares that she has no conflicts of interest.
Human and animal rights
This article does not contain any studies with human participants or animals performed by any of the authors.
Additional information
Edited by Juergen A Richt.
Rights and permissions
About this article
Cite this article
Lawrence, P., Rieder, E. Insights into Jumonji C-domain containing protein 6 (JMJD6): a multifactorial role in foot-and-mouth disease virus replication in cells. Virus Genes 53, 340–351 (2017). https://doi.org/10.1007/s11262-017-1449-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11262-017-1449-8