Abstract
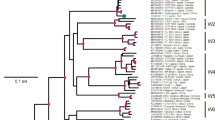
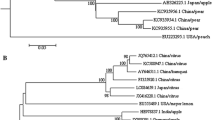
The complete genomic sequences of a Chinese ASPV isolates KL1 and KL9 were determined from ten overlapping cDNA clones. The genomes of both isolates were 9265 nucleotides excluding the poly (A) tail and contained five open reading frames (ORFs). The identities between two complete genomes were 92.5% at nt level. Multiple alignment of the amino acid sequences showed that 110 aa variations between two genomic sequences and the variable domains mainly distributed in 5′-terminal of ORF1, ORF3, and ORF5, respectively. Two complete genomic sequences shared 71.4–77.3% identities with other ASPV isolates at nt level. Phylogenetic relationship analysis of the coat protein genes revealed that ASPV isolates had high variables and formed three groups. All ASPV isolates from apples were clustered to group I, whereas pear were clustered to groups II (except NC_003462) and both KL1 and KL9 were clustered to group III. Nucleotide sequences diversity analysis showed that the between-population d NS /d S ratio 0.092 was similar to these for within-group (0.092–0.095); there was no geographic differentiation between ASPV isolates.

Similar content being viewed by others
References
G.P. Martelli, W. Jelkmann, Arch. Virol. 143, 1245–1249 (1998)
W. Jelkmann, Apple stem pitting virus, in Filamentous viruses of woody plants, ed. by P.L. Monette (Research Signpost, Trivandrum, 1997), pp. 133–142
R.F. Stouffer, Apple stem pitting virus, in Virus and virus-like diseases of pome fruit and simulating noninfectious disorders, ed. by P.L. Fridlund (Washington State University, Pullman, 1989), pp. 138–144
G. Leone, J.L. Lindner, F.A. van der Meer, C.D. Schoen, G. Jongedijk, Acta Hortic. 472, 61–65 (1998)
W. Jelkmann, J. Gen. Virol. 75, 1535–1542 (1994)
H. Koganezawa, H. Yanase, Plant Dis. 74, 610–614 (1990)
N. Galiakparov, D.E. Goszczynski, X. Che, O. Batuman, M. Bar-Joseph, M. Mawassi, Virology 312, 434–448 (2003)
L. Nemchinov, A. Hadidi, F. Faggioli, Acta Hortic. 472, 67–73 (1998)
K. Schwarz, W. Jelkmann, Acta Hortic. 472, 75–85 (1998)
N. Yoshikawa, H. Matsuda, Y. Oda, M. Isogai, T. Takahashi, T. Takahasi, T. Ito, K. Yoshida, Acta Hortic. 550, 285–290 (2001)
F. García-Arenal, A. Fraile, J.M. Malpica, Annu. Rev. Phytopathol. 39, 157–186 (2001)
M. Tsompana, J. Abad, M. Purgganan, W. Moyer, J. Mol. Ecol. 14, 53–66 (2005)
I.M. Moreno, J.M. Malpica, J.A. Díaz-Pendόn, E. Moriones, A. Fraile, F. García-Arenal, Virology 318, 451–460 (2004)
Y. Tomitaka, K. Ohshima, J. Mol. Ecol. 15, 4437–4457 (2006)
R. Biek, A.J. Drummond, M. Poss, Science 311, 538–541 (2006)
M. He, H.S. Li, Y. Zhao, J.X. Niu, B.G. Ma, J. Shihezi Univ. 22, 474–476 (2004)
A.R. Mushegian, E.V. Koonin, Arch. Virol. 133, 239–257 (1993)
P. Pomilo, N.O. Bianchi, Mol. Biol. Evol. 10, 271–281 (1993)
W.H. Li, J. Mol. Ecol. 36, 96–99 (1993)
S.Y. Morozov, A.G. Solovyev, J. Gen. Virol. 84, 1351–1366 (2003)
J. Verchot-Lubicz, Mol. Plant Microbe Interact. 18, 283–290 (2005)
B.Z. Meng, S.Z. Pang, P.L. Forsline, J.R. McFerson, D. Gonsalves, J. Gen. Virol. 79, 2059–2069 (1998)
M.H. Rozanov, E.V. Koonin, A.E. Gorbalenya, J. Gen. Virol. 73, 2129–2134 (1992)
A.E. Gorbalenya, E.V. Koonin, A.P. Donchenko, V.M. Blinov, FEBS Lett. 235, 16–24 (1988)
E.V. Koonin, J. Gen. Virol. 72, 2197–2206 (1991)
A.E. Gorbalenya, V.M. Blinov, A.P. Donchenko, E.V. Koonin, J. Mol. Evol. 28, 256–268 (1989)
V.V. Dolja, V.P. Boyko, A.A. Agranovsky, E.V. Koonin, Virology 184, 79–86 (1991)
L. Giunchedi, C. Poggi Pollini, J. Phytopathol. 134, 329–335 (1992)
W. Jelkmann, L. Kunze, H.J. Vetten, D.E. Lesemann, Acta Hortic. 309, 55–62 (1992)
A. Michel, V. Salmon Marina, J. Plant Pathol. (European) 108, 755–762 (2002)
L.L. Li, Y.F. Dong, Z.P. Zhang, Z.H. Zhang, X.D. Fan, G.Q. Pei, Acta Hortic. Sinica (China) 37, 9–16 (2010)
C.K. Hjulsager, B.S. Olsen, D.M.K. Jensen, M.I. Cordea, B.N. Krath, I.E. Johansen, O.S. Lund, Virology 355, 52–61 (2006)
P. Saénz, M.T. Cervera, S. Dallot, L. Quiot, J.-B. Quiot, J.L. Riechmann, J.A. García, J. Gen. Virol. 81, 557–566 (2000)
V. Decroocq, B. Salvador, O. Sicard, M. Glasa, P. Cosson, L. Svanella-Dumas, F. Revers, J.A. Garcia, T. Candresse, Mol. Plant Microbe Interact. 22, 1311–1320 (2009)
B. Salvador, M.O. Delgadillo, P. Saenz, J.A. Garcia, C. Simon-Mateo, Mol. Plant Microbe Interact. 21, 20–29 (2003)
Acknowledgments
This study was supported by National Natural Science Foundation of China (30360066), the National Key Technologies R&D Program of China (2003BA546C), the Foundation Science and Technology Commission Xinjiang Production and Construction Crops, China (NKB02SDXNK01SW), and Natural Science and Technology Innovation of Shihezi University, China (ZRKX200707).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Liu, N., Niu, J. & Zhao, Y. Complete genomic sequence analyses of Apple stem pitting virus isolates from China. Virus Genes 44, 124–130 (2012). https://doi.org/10.1007/s11262-011-0666-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11262-011-0666-9