Abstract
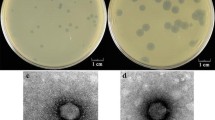
Exponential growing cultures of Streptococcus oralis strain OMZ 1038, isolated from human supragingival dental plaque, were found to release a bacteriophage (designated PH10) upon treatment with mitomycin C. The complete genome sequence of phage PH10 was determined. The genome was 31276 bp in size and contained 54 open reading frames. The module encoding structural proteins was highly similar to that of Streptococcus pneumoniae prophage PhiSpn_3. The most abundant phage structural protein was encoded by ORF35 and was likely processed by proteolytic cleavage. The putative endolysin from PH10, which contained a muramidase domain and a choline-binding domain, was purified and shown to have lytic activity with S. oralis, S. pneumoniae and Streptococcus mitis, but not with other streptococcal species.



Similar content being viewed by others
References
B.J. Paster, S.K. Boches, J.L. Galvin, R.E. Ericson, C.N. Lau, V.A. Levanos, A. Sahasrabudhe, F.E. Dewhirst, Bacterial diversity in human subgingival plaque. J. Bacteriol. 183, 3770–3783 (2001)
C. Pearce, G.H. Bowden, M. Evans, S.P. Fitzsimmons, J. Johnson, M.J. Sheridan, R. Wientzen, M.F. Cole, Identification of pioneer viridans streptococci in the oral cavity of human neonates. J. Med. Microbiol. 42, 67–72 (1995)
M.F. Cole, S. Bryan, M.K. Evans, C.L. Pearce, M.J. Sheridan, P.A. Sura, R. Wientzen, G.H.W. Bowden, Humoral immunity to commensal oral bacteria in human infants: salivary antibodies reactive with Actinomyces naeslundii genospecies 1 and 2 during colonization. Infect. Immun. 66, 4283–4289 (1998)
C.W. Douglas, J. Heath, K.K. Hampton, F.E. Preston, Identity of viridans streptococci isolated from cases of infective endocarditis. J. Med. Microbiol. 39, 179–182 (1993)
L.S. Håvarstein, P. Gaustad, I.F. Nes, D.A. Morrison, Identification of the streptococcal competence-pheromone receptor. Mol. Microbiol. 21, 863–869 (1996)
F. Chi, O. Nolte, C. Bergmann, M. Ip, R. Hakenbeck, Crossing the barrier: evolution and spread of a major class of mosaic pbp2x in Streptococcus pneumoniae, S. mitis and S. oralis. Int. J. Med. Microbiol. 297, 503–512 (2007)
C.A. Tylenda, C. Calvert, P.E. Kolenbrander, A. Tylenda, Isolation of Actinomyces bacteriophage from human dental plaque. Infect. Immun. 49, 1–6 (1985)
A.L. Delisle, C.A. Rostkowski, Lytic bacteriophages of Streptococcus mutans. Curr. Microbiol. 27, 163–167 (1993)
G. Bachrach, M. Leizerovici-Zigmond, A. Zlotkin, R. Naor, D. Steinberg, Bacteriophage isolation from human saliva. Lett. Appl. Microbiol. 36, 50–53 (2003)
G. Hitch, J. Pratten, P.W. Taylor, Isolation of bacteriophages from the oral cavity. Lett. Appl. Microbiol. 39, 215–219 (2004)
R.H. Stevens, B.F. Hammond, C.H. Lai, Characterization of an inducible bacteriophage from a leukotoxic strain of Actinobacillus actinomycetemcomitans. Infect. Immun. 35, 343–349 (1982)
I.R. Siboo, B.A. Bensing, P.M. Sullam, Genomic organization and molecular characterization of SM1, a temperate bacteriophage of Streptococcus mitis. J. Bacteriol. 185, 6968–6975 (2003)
J.R. van der Ploeg, Characterization of Streptococcus gordonii prophage PH15: complete genome sequence and functional analysis of phage-encoded integrase and endolysin. Microbiology 154, 2970–2978 (2008)
H.L. Mitchell, S.G. Dashper, D.V. Catmull, R.A. Paolini, S.M. Cleal, N. Slakeski, K.H. Tan, E.C. Reynolds, Treponema denticola biofilm-induced expression of a bacteriophage, toxin-antitoxin systems and transposases. Microbiology 156, 774–788 (2010)
P. Romero, E. García, T.J. Mitchell, Development of a prophage typing system and analysis of prophage carriage in Streptococcus pneumoniae. Appl. Environ. Microbiol. 75, 1642–1649 (2009)
R. López, E. García, Recent trends on the molecular biology of pneumococcal capsules, lytic enzymes, and bacteriophage. FEMS Microbiol. Rev. 28, 553–580 (2004)
P. Romero, R. López, E. García, Characterization of LytA-like N-acetylmuramoyl-L-alanine amidases from two new Streptococcus mitis bacteriophages provides insights into the properties of the major pneumococcal autolysin. J. Bacteriol. 186, 8229–8239 (2004)
C. Ronda, E. García, R. López, Infection of Streptococcus oralis NCTC 11427 by pneumococcal phages. FEMS Microbiol. Lett. 65, 187–192 (2001)
F.M. Ausubel, R. Brent, R.E. Kingston, D.E. Moore, J.G. Seidman, J.A. Smith, K. Struhl, Current Protocols in Molecular Biology (Wiley, New York, 1987)
X. Huang, A. Madan, CAP3: A DNA sequence assembly program. Genome Res. 9, 868–877 (1999)
A. Lukashin, M. Borodovsky, GeneMark.hmm: new solutions for gene finding. Nucleic Acids Res. 26, 1107–1115 (1998)
M. Kilian, L. Mikkelsen, J. Henrichsen, Taxonomic study of viridans streptococci: description of Streptococcus gordonii sp. nov. and emended descriptions of Streptococcus sanguis (White and Niven 1946), Streptococcus oralis (Bridge and Sneath 1982), and Streptococcus mitis (Andrewes and Horder 1906). Int. J. Syst. Bacteriol. 39, 471–484 (1989)
C. Canchaya, C. Proux, G. Fournous, A. Bruttin, H. Brüssow, Prophage genomics. Microbiol. Mol. Biol. Rev. 67, 238–276 (2003)
P. Romero, N.J. Croucher, N.L. Hiller, F.Z. Hu, G.D. Ehrlich, S.D. Bentley, E. García, T.J. Mitchell, Comparative genomic analysis of 10 Streptococcus pneumoniae temperate bacteriophages. J. Bacteriol. 191, 4854–4862 (2009)
S. Lucchini, F. Desiere, H. Brüssow, Similarly organized lysogeny modules in temperate Siphoviridae from low GC content Gram-positive bacteria. Virology 263, 427–435 (1999)
D. Llull, R. López, E. García, Skl, a novel choline-binding N-acetylmuramoyl-l-alanine amidase of Streptococcus mitis SK137 containing a CHAP domain. FEBS Lett. 580, 1959–1964 (2006)
C. Weigel, H. Seitz, Bacteriophage replication modules. FEMS Microbiol. Rev. 30, 321–381 (2006)
M. Zuniga, B. Franke-Fayard, G. Venema, J. Kok, A. Nauta, Characterization of the putative replisome organizer of the lactococcal bacteriophage r1t. J. Virol. 76, 10234–10244 (2002)
V. Obregón, P. García, R. López, J.L. García, VO1, a temperate bacteriophage of the type 19A multiresistant epidemic 8249 strain of Streptococcus pneumoniae: analysis of variability of lytic and putative C5 methyltransferase genes. Microb. Drug Resist. 9, 7–15 (2003)
M. Radlińska, A. Piekarowicz, Cloning and characterization of the gene encoding a new DNA methyltransferase from Neisseria gonorrhoeae. Biol. Chem. 379, 1391–1395 (1998)
M. Radlińska, J.M. Bujnicki, A. Piekarowicz, Structural characterization of two tandemly arranged DNA methyltransferase genes from Neisseria gonorrhoeae MS11: N4-cytosine specific M.NgoMXV and nonfunctional 5-cytosine-type M. NgoMorf2P. Proteins 37, 717–728 (1999)
A. van Belkum, S. Scherer, L. van Alphen, H. Verbrugh, Short-sequence DNA repeats in prokaryotic genomes. Microbiol. Mol. Biol. Rev. 62, 275–293 (1998)
G.T. Chung, J.S. Yoo, H.B. Oh, Y.S. Lee, S.H. Cha, S.J. Kim, C.K. Yoo, Complete genome sequence of Neisseria gonorrhoeae NCCP11945. J. Bacteriol. 190, 6035–6036 (2008)
E. García, J.L. García, P. García, A. Arrarás, J.M. Sánchez-Puelles, R. López, Molecular evolution of lytic enzymes of Streptococcus pneumoniae and its bacteriophages. Proc. Natl. Acad. Sci. USA 85, 914–918 (1988)
J.A. Hermoso, B. Monterroso, A. Albert, B. Galan, O. Ahrazem, P. García, M. Martinez-Ripoll, J.L. García, M. Menéndez, Structural basis for selective recognition of pneumococcal cell wall by modular endolysin from phage Cp-1. Structure 11, 1239–1249 (2003)
I. Pérez-Dorado, N.E. Campillo, B. Monterroso, D. Hesek, M. Lee, J.A. Páez, P. García, M. Martínez-Ripoll, J.L. García, S. Mobashery, M. Menéndez, J.A. Hermoso, Elucidation of the molecular recognition of bacterial cell wall by modular pneumococcal phage endolysin Cpl-1. J Biol. Chem. 34, 24990–24999 (2007)
Acknowledgments
The excellent technical assistance of Verena Osterwalder and Steve Reese is gratefully acknowledged. I thank Peter Brunisholz and Serge Chesnov for mass spectrometry analysis.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
van der Ploeg, J.R. Genome sequence of the temperate bacteriophage PH10 from Streptococcus oralis . Virus Genes 41, 450–458 (2010). https://doi.org/10.1007/s11262-010-0525-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11262-010-0525-0