Abstract
Paratuberculosis (PTB) is a chronic infectious enteritis of ruminants, caused by Mycobacterium avium subspecies paratuberculosis (MAP) that brings huge economic loss to the dairy farmers. The study was conducted to explore the association of selected SNPs in IFNG, SLC11A1, ANKRA2 and PGLYRP1 genes with resistance to PTB disease in Indian cattle population. A case-control resource population was established based on the results of diagnostic tests used for detection of MAP infection status viz. ELISA, Johnin PPD test, faecal microscopy and IS900 blood PCR. The PCR-RFLP method was used for genotyping of SNPs. SNPs rs109453173 in SLC11A1, rs110853455 in IFNG and rs41933863 in ANKRA2 genes were significantly (P<0.05) associated with resistance to MAP infection. For SNP rs109453173, GG genotype and G allele was found to be associated with resistance against MAP infection than CC and CG genotypes and C allele, respectively. For SNP rs110853455, AG genotype was found to be associated with susceptibility to MAP infection than AA and GG genotype. For SNP rs41933863, the AG genotype provided three and six times more resistance against MAP infection than GG and AA genotype. The results of this study are suggestive of SNPs rs109453173, rs110853455 and rs41933863 as potential markers for screening MAP resistant cattle and a breeding programme favouring GG genotype and G allele for rs109453173, AG genotype for rs41933863 and against AG genotype for rs110853455 might confer resistance against MAP infection in Indian cattle. However, investigation of these SNPs in an independent and larger population will warrant the strength of association for resistance against MAP infection in cattle.
Similar content being viewed by others
Data availability
The data will be available on request to the corresponding author.
Code availability
Not applicable.
References
Abraham A, Naicy T, Raghavan KC, Siju J, Aravindakshan T (2017) Evaluation of the association of SLC11A1 gene polymorphism with incidence of paratuberculosis in goats. J Genet 96(4):641–646. https://doi.org/10.1007/s12041-017-0820-9
Awomoyi AA (2007) The human solute carrier family 11 member 1 protein (SLC11A1): Linking infections, autoimmunity and cancer? FEMS Immunol Med Microbiol 49(3):324–329. https://doi.org/10.1111/j.1574-695X.2007.00231.x
Baqir M, Bhushan S, Kumar A et al (2016) Association of polymorphisms in SLC11A1 gene with bovine tuberculosis trait among Indian cattle. J Appl Anim Res 44(1):380–383. https://doi.org/10.1080/09712119.2015.1091333
Bastida F, Juste RA (2011) Paratuberculosis control: A review with a focus on vaccination. J Immune Based Ther Vaccines 9(1):1–7. https://doi.org/10.1186/1476-8518-9-8
Beard PM, Daniels MJ, Henderson D, Pirie A, Rudge K, Buxton D, Rhind S, Greig A, Hutchings MR, McKendrick I, Stevenson K (2001a) Paratuberculosis infection of nonruminant wildlife in Scotland. J Clin Microbiol 39(4):1517–1521. https://doi.org/10.1128/FJCM.39.4.1517-1521.2001
Beard PM, Rhind SM, Buxton D, Daniels MJ, Henderson D, Pirie A, Rudge K, Greig A, Hutchings MR, Stevenson K, Sharp JM (2001b) Natural paratuberculosis infection in rabbits in Scotland. J Comp Pathol 124(4):290–299. https://doi.org/10.1053/jcpa.2001.0466
Bhutediya JM, Dandapat P, Chakrabarty A, Das R, Nanda PK, Bandyopadhyay S, Biswas TK (2017) Prevalence of paratuberculosis in organized and unorganized dairy cattle herds in West Bengal, India. Vet World 10(6):574–579. https://doi.org/10.14202/vetworld.2017.574-579
Bishop SC, MacKenzie KM (2003) Genetic management strategies for controlling infectious diseases in livestock populations. Genet Sel Evol 35(1):1–5. https://doi.org/10.1186/1297-9686-35-s1-s3
Bressani FA, Tizioto PC, Giglioti R et al (2014) Single nucleotide polymorphisms in candidate genes associated with gastrointestinal nematode infection in goats. Genet Mol Res 13(4):8530–8536. https://doi.org/10.4238/2014.October.20.29
Canonne-Hergaux F, Gruenheid S, Govoni G, Gros P (1999) The Nramp1 protein and its role in resistance to infection and macrophage function. Proc Assoc Am Physicians 111(4):283-289. https://doi.org/10.1046/j.1525-1381.1999.99236.x
Casas E, Garcia MD, Wells JE, Smith TPL (2011) Association of single nucleotide polymorphisms in the ANKRA2 and CD180 genes with bovine respiratory disease and presence of Mycobacterium avium subsp. paratuberculosis. Anim Genet 42(6):571–577. https://doi.org/10.1111/j.1365-2052.2011.02189.x
Chase CCL, Hurley DJ, Reber AJ (2008) Neonatal immune development in the calf and its impact on vaccine response. Vet Clin North Am - Food Anim Pract 24(1):87–104. https://doi.org/10.1016/j.cvfa.2007.11.001
Chiodini RJ, Van Kruiningen HJ, Merkal RS (1984) Ruminant paratuberculosis (Johne’s disease): the current status and future prospects. Cornell Vet 74:218–262
Cossu D, Masala S, Cocco E, Paccagnini D, Tranquilli S, Frau J, Marrosu MG, Sechi LA (2013) Association of Mycobacterium avium subsp. paratuberculosis and SLC11A1 polymorphisms in Sardinian multiple sclerosis patients. J Infect Dev Ctries 7(03):203-207. https://doi.org/10.3855/jidc.2737
Coussens PM, Verman N, Coussens MA, Elftman MD, McNulty AM (2004) Cytokine gene expression in peripheral blood mononuclear cells and tissues of cattle infected with Mycobacterium avium subsp. paratuberculosis: Evidence for an inherent proinflammatory gene expression pattern. Infect Immun 72(3):1409–1422. https://doi.org/10.1128/IAI.72.3.1409-1422.2004
Dervishi E, Uriarte J, Valderrábano J, Calvo JH (2011) Structural and functional characterisation of the ovine interferon gamma (IFNG) gene: Its role in nematode resistance in Rasa Aragonesa ewes. Vet Immunol Immunopathol 141(1-2):100–108. https://doi.org/10.1016/j.vetimm.2011.02.013
Ding X, Zhang X, Yang Y, Ding Y, Xue W, Meng Y, Zhu W, Yin Z (2014) Polymorphism, expression of natural resistance-associated macrophage protein 1 encoding gene (NRAMP1) and its association with immune traits in pigs. Asian-Australasian J Anim Sci 27(8):1189–1195. https://doi.org/10.5713/ajas.2014.14017
Ellingson JLE, Anderson JL, Koziczkowski JJ, Radcliff RP, Sloan SJ, Allen SE, Sullivan NM (2005) Detection of viable Mycobacterium avium subsp. paratuberculosis in retail pasteurized whole milk by two culture methods and PCR. J Food Prot 68(5):966–972. https://doi.org/10.4315/0362-028X-68.5.966
Fodil M, Teixeira VH, Chaudru V, Hilliquin P, Bombardieri S, Balsa A, Westhovens R, Barrera P, Alves H, Migliorin P, Bardin T (2015) Relationship between SNPs and expression level for candidate genes in rheumatoid arthritis. Scand J Rheumatol 44(1):2–7. https://doi.org/10.3109/03009742.2014.918175
Fox NJ, Smith LA, Stevenson K, Davidson RS, Marion G, Hutchings MR (2020) Infection of non-ruminant wildlife by Mycobacterium avium subsp. paratuberculosis. In Behr MA, Stevenson K, Kapur V (eds). Paratuberculosis 2020:200-212
Ganguly I, Sharma A, Mitra A, Kumar N, Ganguly A (2011) Analysis of genetic variations of complete tm4 of buffalo (Bubalus bubalis) SLC11A1 gene. J Appl Anim Res 39(4):324–327. https://doi.org/10.1080/09712119.2011.607943
Garcia AB, Shalloo L (2015) Invited review: The economic impact and control of paratuberculosis in cattle. J Dairy Sci 98(8):5019–5039. https://doi.org/10.3168/jds.2014-9241
Garg R, Patil PK, Singh SV, Sharma S, Gandham RK, Singh AV, Filia G, Singh PK, Jayaraman S, Gupta S, Chaubey KK (2015) Comparative evaluation of different test combinations for diagnosis of Mycobacterium avium subspecies paratuberculosis infecting dairy herds in India. Biomed Res Int :Article ID 983978. https://doi.org/10.1155/2015/983978
Gonda MG, Chang YM, Shook GE, Collins MT, Kirkpatrick BW (2006) Genetic variation of Mycobacterium avium ssp. paratuberculosis infection in US Holsteins. J Dairy Sci 89(5):1804–1812. https://doi.org/10.3168/jds.S0022-0302(06)72249-4
Gonsky R, Deem RL, Landers CJ, Haritunians T, Yang S, Targan SR (2014) IFNG rs1861494 polymorphism is associated with IBD disease severity and functional changes in both IFNG methylation and protein secretion. Inflamm Bowel Dis 20(10):1794-1801. https://doi.org/10.1097/MIB.0000000000000172
Gopi B, Singh RV, Kumar S, Kumar S, Chauhan A, Kumar A, Singh SV (2020) Single-nucleotide polymorphisms in CLEC7A, CD209 and TLR4 gene and their association with susceptibility to paratuberculosis in Indian cattle. J Genet 99(1):1–10. https://doi.org/10.1007/s12041-019-1172-4
Gupta A, Rani M, Agrawal S, Kumar Gupta P (2012) Sero-prevalence of paratuberculosis (Johne’s Disease) in cattle population of south-western bangalore using ELISA kit. Open J Vet Med 02(4):196–200. https://doi.org/10.4236/ojvm.2012.24031
Halbert ND, Cohen ND, Slovis NM, Faircloth J, Martens RJ (2006) Variations in equid SLC11A1 (NRAMP1) genes and associations with Rhodococcus equi pneumonia in horses. J Vet Intern Med 20(4):974–979. https://doi.org/10.1892/0891-6640(2006)20[974:VIESNG]2.0.CO;2
Hinger M, Brandt H, Horner S, Erhard G (2007) Short communication: Association analysis of microsatellites and Mycobacterium avium subspecies paratuberculosis antibody response in German holsteins. J Dairy Sci 90(4):1957–1961. https://doi.org/10.3168/jds.2006-510
Holder A, Garty R, Elder C, Mesnard P, Laquerbe C, Bartens MC, Salavati M, Shabbir MZ, Tzelos T, Connelly T, Villarreal-Ramos B (2020) Analysis of genetic variation in the bovine SLC11A1 Gene, Its influence on the expression of NRAMP1 and potential association with resistance to bovine tuberculosis. Front Microbiol 11:1420. https://doi.org/10.3389/fmicb.2020.01420
Hruska K, Bartos M, Kralik P, Pavlik I (2005) Mycobacterium avium subsp. paratuberculosis in powdered infant milk: paratuberculosis in cattle-the public health problem to be solved. Vet Med (Praha) 50(8):327–335
Hruska K, Slana I, Kralik P, Pavlik I (2011) Mycobacterium avium subsp. paratuberculosis in powdered infant milk: F57 competitive real time PCR. Vet Med (Praha) 56(5):226–230
Iannaccone M, Cosenza G, Pauciullo A, Martino G, Capparelli R (2018) The SNP g.4667G>A at 3′-UTR of IFNG gene is associated with susceptibility to bovine tuberculosis in Mediterranean water buffalo (Bubalus bubalis). Anim Genet 49(5):496–497. https://doi.org/10.1111/age.12698
Juste RA, Perez V (2011) Control of paratuberculosis in sheep and goats. Vet Clin North Am - Food Anim Pract 27(1):127–138. https://doi.org/10.1016/j.cvfa.2010.10.020
Juste RA, Vazquez P, Ruiz-Larrañaga O, Iriondo M, Manzano C, Agirre M, Estonba A, Geijo MV, Molina E, Sevilla IA, Alonso-Hearn M (2018) Association between combinations of genetic polymorphisms and epidemiopathogenic forms of bovine paratuberculosis. Heliyon 4(2):e00535. https://doi.org/10.1016/j.heliyon.2018.e00535
Krawczyk M, Masternak K, Zufferey M, Barras E, Reith W (2005) New functions of the major histocompatibility complex class II-specific transcription factor RFXANK revealed by a high-resolution mutagenesis study. Mol Cell Biol 25(19):8607–8618. https://doi.org/10.1128/mcb.25.19.8607-8618.2005
Kumar S (2015) Single nucleotide polymorphism in candidate genes and their association with occurrence of paratuberculosis in cattle. IVRI (Deemed University), Izatnagar Bareilly, UP, India. Accessed on 20-03-2021. http://krishikosh.egranth.ac.in/handle/1/5810096974
Kumar S, Kumar S, Singh R, Chauhan A, Agrawal S, Kumar A, Singh S (2017) Investigation of genetic association of single nucleotide polymorphisms in SP110 gene with occurrence of paratuberculosis disease in cattle. Int J Livest Res 7:81–88. https://doi.org/10.5455/ijlr.20170223021951
Kumar S, Kumar S, Singh RV, Chauhan A, Kumar A, Bharati J, Singh SV (2020) Association of genetic variability in CD209 gene with bovine paratuberculosis disease: a case–control study in the Indian cattle population. Anim Biotechnol 25:1–8. https://doi.org/10.1080/10495398.2020.1823400
Kumar S, Kumar S, Singh RV, Chauhan A, Kumar A, Bharati J, Singh SV (2019a) Association of Bovine CLEC7A gene polymorphism with host susceptibility to paratuberculosis disease in Indian cattle. Res Vet Sci 123:216–222. https://doi.org/10.1016/j.rvsc.2019.01.016
Kumar S, Kumar S, Singh RV, Chauhan A, Kumar A, Sulabh S, Bharati J, Singh SV (2019b) Genetic association of polymorphisms in bovine TLR2 and TLR4 genes with Mycobacterium avium subspecies paratuberculosis infection in Indian cattle population. Vet Res Commun 43(2):105–114. https://doi.org/10.1007/s11259-019-09750-2
Larsen AB, Moon HW, Merkal RS (1971) Susceptibility of swine to Mycobacterium paratuberculosis. Am J Vet Res 32(4):589–595
Lo D, Tynan W, Dickerson J, Mendy J, Chang HW, Scharf M, Byrne D, Brayden D, Higgins L, Evans C, O’Mahony DJ (2003) Peptidoglycan recognition protein expression in mouse Peyer’s Patch follicle associated epithelium suggests functional specialization. Cell Immunol 224(1):8–16. https://doi.org/10.1016/S0008-8749(03)00155-2
Losinger WC (2005) Economic impact of reduced milk production associated with Johne’s disease on dairy operations in the USA. J Dairy Res 72(4):425–432. https://doi.org/10.1017/S0022029905001007
Maryam J, Babar ME, Nadeem A, Hussain T (2012) Genetic variants in interferon gamma (IFN-c) gene are associated with resistance against ticks in Bos taurus and Bos indicus. Mol Biol Rep 39(4):4565–4570. https://doi.org/10.1007/s11033-011-1246-8
Masternak K, Barras E, Zufferey M, Conrad B, Corthals G, Aebersold R, Sanchez JC, Hochstrasser DF, Mach B, Reith WA (1998) A gene encoding a novel RFX-associated transactivator is mutated in the majority of MHC class II deficiency patients. Nat Genet 20(3):273–277. https://doi.org/10.1038/3081
Miller MA, Davey SC, van Helden LS, Kettner F, Weltan SM, Last R, Grewar JD, Botha L, Van Helden PD (2017) Paratuberculosis in a domestic dog in South Africa. J S Afr Vet Assoc 88(1):1–5. https://doi.org/10.4102/jsava.v88i0.1441
Narnaware SD, Tripathi BN (2017) Seroepidemiology of paratuberculosis in cattle population of organized and unorganized farms of India. Indian J Anim Sci 87(1):21–24. https://www.researchgate.net/profile/Shirish-Narnaware/publication/313859367_Seroepidemiology_of_paratuberculosis_in_cattle_population_of_organized_and_unorganized_farms_of_India/links/58abcfb2aca27206d9bd1b24/Seroepidemiology-of-paratuberculosis-in-cattle-population-of-organized-and-unorganized-farms-of-India.pdf
Nielsen SS, Ersbøll AK (2006) Age at occurrence of Mycobacterium avium subspecies paratuberculosis in naturally infected dairy cows. J Dairy Sci 89(12):4557–4566
Nielsen SS, Toft N (2008) Ante mortem diagnosis of paratuberculosis: A review of accuracies of ELISA, interferon-γ assay and faecal culture techniques. Vet Microbiol 129(3-4):217–235. https://doi.org/10.1016/j.vetmic.2007.12.011
OIE (2020) World organisation for animal health. https://www.oie.int/en/animal-health-in-the-world/animal-diseases/PTB/. Accessed 11-06-2020
Onoyama S, Ihara K, Yamaguchi Y, Ikeda K, Yamaguchi K, Yamamura K, Hoshina T, Mizuno Y, Hara T (2012) Genetic susceptibility to Kawasaki disease: Analysis of pattern recognition receptor genes. Hum Immunol 73(6):654–660. https://doi.org/10.1016/j.humimm.2012.03.011
Paixão TA, Martinez R, Santos RL (2012) Polymorphisms of the coding region of SLC11A1 (Nramp1) gene associated to natural resistance against bovine brucellosis. Arq Bras Med Vet e Zootec 64(4):1081–1084. https://doi.org/10.1590/S0102-09352012000400041
Pant SD, Verschoor CP, Schenkel FS, You Q, Kelton DF, Karrow NA (2011a) Bovine PGLYRP1 polymorphisms and their association with resistance to Mycobacterium avium ssp. Paratuberculosis. Anim Genet 42(4):354–360. https://doi.org/10.1111/j.1365-2052.2010.02153.x
Pant SD, Verschoor CP, Skelding AM, Schenkel FS, You Q, Biggar GA, Kelton DF, Karrow NA (2011b) Bovine IFNGR2, IL12RB1, IL12RB2, and IL23R polymorphisms and MAP infection status. Mamm Genome 22(9):583–588. https://doi.org/10.1007/s00335-011-9332-8
Pavlik I, Jahn P, Dvorska L, Bartos M, Novotny L, Halouzka R (2004) Mycobacterial infections in horses: a review of the literature. Vet Med 49(11):427. https://doi.org/10.17221/5733-VETMED
Pinedo PJ, Buergelt CD, Donovan GA, Melendez P, Morel L, Wu R, Langaee TY, Rae DO (2009a) Association between CARD15/NOD2 gene polymorphisms and paratuberculosis infection in cattle. Vet Microbiol 134(3-4):346–352. https://doi.org/10.1016/j.vetmic.2008.09.052
Pinedo PJ, Buergelt CD, Donovan GA, Melendez P, Morel L, Wu R, Langaee TY, Rae DO (2009b) Candidate gene polymorphisms (BoIFNG, TLR4, SLC11A1) as risk factors for paratuberculosis infection in cattle. Prev Vet Med 91(2-4):189-196. https://doi.org/10.1016/j.prevetmed.2009.05.020
Pithua P, Godden SM, Wells SJ, Stabel JR (2011) Evaluation of the risk of paratuberculosis in adult cows fed Mycobacterium avium subsp paratuberculosis DNA-positive or-negative colostrum as calves. Am J Vet Res 72(11):1456–1464. https://doi.org/10.2460/ajvr.72.11.1456
Prakash O, Kumar A, Sonwane A, Rathore R, Singh RV, Chauhan A, Kumar P, Renjith R, Yadav R, Bhaladhare A, Baqir M (2014) Polymorphism of cytokine and innate immunity genes associated with bovine brucellosis in cattle. Mol Biol Rep 41(5):2815–2825. https://doi.org/10.1007/s11033-014-3136-3
Pritchard TC, Coffey MP, Bond KS, Hutchings MR, Wall E (2017) Phenotypic effects of subclinical paratuberculosis (Johne’s disease) in dairy cattle. J Dairy Sci 100(1):679–690. https://doi.org/10.3168/jds.2016-11323
Radostits OM, Gay CC, Hinchcliff KW, Constable PD (eds) (2006) Veterinary Medicine E-Book: A textbook of the diseases of cattle, horses, sheep, pigs and goats. Elsevier Health Sciences, Amsterdam
Rawat KD, Chaudhary S, Kumar N, Gupta S, Chaubey KK, Singh SV, Dhama K, Deb R (2014) Economic losses in a commercial dairy farm due to the outbreak of Johne’s disease in India. Res J Vet Pract 2:73–77. https://doi.org/10.14737/journal.rjvp/2.5.73.77
Reddacliff LA, Beh K, McGregor H, Whittington RJ (2005) A preliminary study of possible genetic influences on the susceptibility of sheep to Johne’s disease. Aust Vet J 83(7):435–441. https://doi.org/10.1111/j.1751-0813.2005.tb13087.x
Rieseberg LH, Archer MA, Wayne RK (1999) Transgressive segregation, adaptation and speciation. Heredity 83(4):363–372
Roupie V, Rosseels V, Piersoel V, Zinniel DK, Barletta RG, Huygen K (2008) Genetic resistance of mice to Mycobacterium paratuberculosis is influenced by SLC11A1 at the early but not at the late stage of infection. Infect Immun 76(5):2099–2105. https://doi.org/10.1128/IAI.01137-07
Ruiz-Larrañaga O, Garrido JM, Manzano C, Iriondo M, Molina E, Gil A, Koets AP, Rutten VP, Juste RA, Estonba A (2010) Identification of single nucleotide polymorphisms in the bovine solute carrier family 11 member 1 (SLC11A1) gene and their association with infection by Mycobacterium avium subspecies paratuberculosis. J Dairy Sci 93(4):1713–1721
Sablik P, Klenowicz A, Szewczuk M, Olszewski A, Dybus A (2020) The Effect of Polymorphism in PGLYRP1 Gene on the Productivity and Health Traits in Holstein-Friesian Cattle. Russ J Genet 56:333–338. https://doi.org/10.1134/S1022795420030138
Sadana T, Singh RV, Singh SV, Saxena VK, Sharma D, Singh PK, Kumar N, Gupta S, Chaubey KK, Jayaraman S, Tiwari R, Dhama K, Bhatia AK, Sohal JS (2015) Single nucleotide polymorphism of SLC11A1, CARD15, IFNG and TLR2 genes and their association with Mycobacterium avium subspecies paratuberculosis infection in native Indian cattle population. Indian J Biotechnol 14:469-475. Accessed on 20-03-2021. http://nopr.niscair.res.in/bitstream/123456789/33994/1/IJBT%2014(4)%20469-475.pdf
Sanchez-Robert E, Altet L, Utzet-Sadurni M, Giger U, Sanchez A, Francino O (2008) SLC11A1 (formerly Nramp1) and susceptibility to canine visceral leishmaniasis. Vet Res 39(3):36 https://doi.org/10.1051/vetres:2008013
Sechi LA, Dow CT (2015) Mycobacterium avium ss. paratuberculosis Zoonosis - The Hundred Year War - Beyond Crohn’s Disease. Front Immunol 6:96. https://doi.org/10.3389/fimmu.2015.00096
Sechi LA, Gazouli M, Sieswerda LE, Molicotti P, Ahmed N, Ikonomopoulos J, Scanu AM, Paccagnini D, Zanetti S (2006) Relationship between Crohn’s disease, infection with Mycobacterium avium subspecies paratuberculosis and SLC11A1 gene polymorphisms in Sardinian patients. World J Gastroenterol. 12(44):7161-7164. https://doi.org/10.3748/wjg.v12.i44.7161
Shankar H, Singh SV, Singh PK, Singh AV, Sohal JS, Greenstein RJ (2010) Presence, characterization, and genotype profiles of Mycobacterium avium subspecies paratuberculosis from unpasteurized individual and pooled milk, commercial pasteurized milk, and milk products in India by culture, PCR, and PCR-REA methods. Int J Infect Dis 14(2):e121–e126. https://doi.org/10.1016/j.ijid.2009.03.031
Shivashanker JM, Nagaraja R, Yathiraj S, Rajeshwari YB, Isloor S (2018) Genetic Polymorphism of PGLYRP-1 Gene and its Association with Mastitis in Deoni and Holstein Friesian Crossbred Cows. Rumin Sci 7:199–202. Accessed on 20-03-2021. https://www.anandpub.com/archive-june-volume-2018-7-1-issue6/
Shtrichman R, Samuel CE (2001) The role of gamma interferon in antimicrobial immunity. Curr Opin Microbiol 4(3):251–259. https://doi.org/10.1016/S1369-5274(00)00199-5
Singh SV, Singh PK, Gupta S, Chaubey KK, Singh B, Kumar A, Singh AV, Kumar N (2013) Comparison of microscopy and blood-PCR for the diagnosis of clinical Johne’s disease in domestic ruminants. Iran J Vet Res 14(4):345-349. Accessed on 20-03-2021. https://ijvr.shirazu.ac.ir/article_1833_d43044a2bd892c904f91ef1040a56ace.pdf
Singh SV, Singh AV, Singh R, Sharma S, Shukla N, Misra S, Singh PK, Sohal JS, Kumar H, Patil PK, Misra P (2008) Sero-prevalence of Bovine Johne’s disease in buffaloes and cattle population of North India using indigenous ELISA kit based on native Mycobacterium avium subspecies paratuberculosis “Bison type” genotype of goat origin. Comp Immunol Microbiol Infect Dis 31(5):419–433. https://doi.org/10.1016/j.cimid.2007.06.002
Stewart LC, Day AS, Pearson J, Barclay ML, Gearry RB, Roberts RL, Bentley RW (2010) SLC11A1 polymorphisms in inflammatory bowel disease and Mycobacterium avium subspecies paratuberculosis status. World J Gastroenterol 16(45):5727–5731. https://doi.org/10.3748/wjg.v16.i45.5727
Tydell CC, Yuan J, Tran P, Selsted ME (2006) Bovine peptidoglycan recognition protein-S: Antimicrobial activity, localization, secretion, and binding properties. J Immunol 176(2):1154–1162. https://doi.org/10.4049/jimmunol.176.2.1154
Vázquez P, Ruiz-Larrañaga O, Garrido JM, Iriondo M, Manzano C, Agirre M, Estonba A, Juste RA (2014) Genetic association analysis of paratuberculosis forms in Holstein-Friesian cattle. Vet Med Int Article ID 321327. https://doi.org/10.1155/2014/321327
Vincze T, Posfai J, Roberts RJ (2003) NEBcutter: A program to cleave DNA with restriction enzymes. Nucleic Acids Res 31(13):3688–3691. https://doi.org/10.1093/nar/gkg526
Wang HL, Li ZX, Wang LJ, He H, Yang J, Chen L, Niu FB, Liu Y, Guo JZ, Liu XL (2013) Polymorphism in PGLYRP-1 gene by PCR-RFLP and its association with somatic cell score in Chinese Holstein. Res Vet Sci 95(2):508–514. https://doi.org/10.1016/j.rvsc.2013.06.005
Whittington R, Donat K, Weber MF, Kelton D, Nielsen SS, Eisenberg S, Arrigoni N, Juste R, Sáez JL, Dhand N, Santi A, … De Waard JH (2019) Control of paratuberculosis: Who, why and how. A review of 48 countries. BMC Vet Res 15(1):1–29. https://doi.org/10.1186/s12917-019-1943-4
Whittington R, Tizard M, Reddacliff L (2004) Mycobacterium paratuberculosis: pain in the gut and challenge for science. Microbiol Aust 25(4):36–37. https://doi.org/10.1071/ma04436
Wu S, Liu X, Wang Y, Zhang M, Wang M, He JQ (2019) Genetic polymorphisms of IFNG and IFNGr1 with latent tuberculosis infection. Dis Markers 2019. Article ID 8410290 https://doi.org/10.1155/2019/8410290
Yadav R, Sharma AK, Singh R, Sonwane AR, Kumar A, Chauhan A, Kumar S, Kumar T, Renjith R, Bhaladhare A, Prakash O (2014) An association study of snps with susceptibility to bovine paratuberculosis infection in cattle. Indian J Anim Sci 84(5):490–493
Zulfiqar F, Hozo I, Rangarajan S, Mariuzza RA, Dziarski R, Gupta D (2013) Genetic association of peptidoglycan recognition protein variants with inflammatory bowel disease. PLoS One 8(6):e67393. https://doi.org/10.1371/journal.pone.0067393
Acknowledgements
Authors are thankful to Director, ICAR-Indian Veterinary Research Institute, Izatnagar, India for providing necessary facilities for conducting this work.
Funding
The work was supported by the Director, ICAR-Indian Veterinary Research Institute, Izatnagar, India for providing necessary facilities and funding of this work with grant no. IXX09774 to Dr. Ran Vir Singh.
Author information
Authors and Affiliations
Contributions
RVS, SAK and AC conceived and designed the study and provided resources. BG, SAK, SUK and AC screened the animals and performed genotyping experiments. BG and SVS performed IS900 blood PCR. AK, JB and SAK analysed the data. BG and SAK wrote the original draft. SAK and JB reviewed and edited the original draft. All authors discussed the results, edited the manuscript and approved the final version of the manuscript for publication.
Corresponding authors
Ethics declarations
Conflicts of interest/Competing interests
The authors do not have any conflict of interest.
Ethics approval
All the animal experimentation had prior approval of the Institutional Animal Ethics Committee (ICAR-Indian Veterinary Research Institute, Izatnagar, India), which follows the guidelines of CPCSEA (Committee for the Purpose of Control and Supervision of Experimentation in Animals), Government of India. The approval of IAEC was given for the experiments conducted in the project vide IXX09774.
Consent to participate
Not applicable.
Consent for publication
All authors had approved the final version of the manuscript for publication.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Supplementary material Figure 1
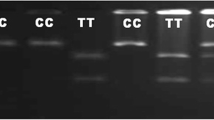
Genotypic profile of SNP rs109453173 in SLC11A1 gene by PCR-RFLP with 100-bp marker (M represents marker lane). (DOCX 161 kb)
Supplementary material Figure 2
Genotypic profile of SNP rs41933863 in ANKRA2 gene by PCR-RFLP with 100-bp marker (M represents marker lane). (DOCX 68 kb)
Supplementary material Table 1
(DOCX 16 kb)
Supplementary material Table 2
(DOCX 15 kb)
Supplementary material Table 3
(DOCX 13 kb)
Rights and permissions
About this article
Cite this article
Gopi, B., Vir Singh, R., Kumar, S. et al. Effect of selected single nucleotide polymorphisms in SLC11A1, ANKRA2, IFNG and PGLYRP1 genes on host susceptibility to Mycobacterium avium subspecies paratuberculosis infection in Indian cattle. Vet Res Commun 46, 209–221 (2022). https://doi.org/10.1007/s11259-021-09849-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11259-021-09849-5