Abstract
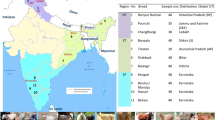
Large-scale mitochondrial DNA (mtDNA) D-loop sequences data from previous studies were investigated to obtain genetic information which contributes to a better understanding of the genetic diversity and history of modern sheep. In this study, we analyzed mtDNA D-loop sequences of 963 individuals from 16 Chinese indigenous breeds that distributed seven geographic regions. Phylogenetic analysis showed that all three previously defined haplogroups A, B, and C were found in all breeds among different regions except in Southwest China mountainous region, which had only the A and B haplogroups. The weak phylogeographic structure was observed among Chinese indigenous sheep breeds distribution regions and this could be attributable to long-term strong gene flow among regions induced by the human migration, commercial trade, and extensive transport of sheep. The estimation of demographic parameters from mismatch analyses showed that haplogroups A and B had at least one demographic expansion of indigenous sheep in China.





Similar content being viewed by others
Abbreviations
- mtDNA:
-
mitochondrial DNA
References
Chen, S.Y., Duan, Z.Y., Sha, T., Xiangyu, J., Wu, S.F., Zhang, Y.P., 2006. Origin, genetic diversity, and population structure of Chinese domestic sheep. Gene 376: 216–223
Excoffier, L., Laval, G., Schneider, S., 2005. Arlequin (version 3.0): an integrated software package for population genetics data analysis. Evolutionary Bioinformatics Online, 1: 47
Excoffier, L., Smouse, P.E., Quattro, J.M., 1992. Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data. Genetics, 131(2): 479–491.
Fu, Y.-X., 1997. Statistical tests of neutrality of mutations against population growth, hitchhiking and background selection. Genetics, 147(2): 915–925.
Guan, S., He, X.-h., Ya-bin, P., Ye, S.-h., Guan, W.-j., Ma, Y.-h., 2007. Study on mtDNA genetic diversity and phylogeny evolution of 5 indigenous sheep populations in southwest China. Acta Veterinaria et Zootechnica Sinica 38, 219–224
Guo, J., Du, L.X., Ma, Y.H., Guan, W.J., Li, H.B., Zhao, Q.J., Li, X., Rao, S.Q., 2005. A novel maternal lineage revealed in sheep (Ovis aries). Animal Genetics 36, 331–336. doi:10.1111/j.1365-2052.2005.01310.x
Hiendleder, S., Mainz, K., Plante, Y., Lewalski, H., 1998. Analysis of mitochondrial DNA indicates that domestic sheep are derived from two different ancestral maternal sources: no evidence for contributions from Urial and Argali sheep. Journal of Heredity, 89(2): 113–120.
Huelsenbeck, J.P., Ronquist, F., 2001. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics, 17(8): 754–755.
Kimura, M., 1980. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. Journal of Molecular Evolution, 16(2): 111–120.
Larkin, M. et al., 2007. Clustal W and Clustal X version 2.0. Bioinformatics, 23(21): 2947–2948.
Librado, P., Rozas, J., 2009. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics, 25(11): 1451–1452.
Luo, Y.-Z., Cheng, S.-R., Lkhagva, B., Badamdorj, D., Hanotte, O., Han, J.-L., 2005. Origin and genetic diversity of Mongolian and Chinese sheep using mitochondrial DNA D-loop sequences. Acta Genetica Sinica 32, 1256–1265.
Naderi, S., Rezaei, H.-R., Taberlet, P., Zundel, S., Rafat, S.-A., Naghash, H.-R., El-Barody, M.A.A., Ertugrul, O., Pompanon, F., Econogene, C., 2007. Large-scale mitochondrial DNA analysis of the domestic goat reveals six haplogroups with high diversity. Plos One 2(10): e1012. doi:10.1371/journal.pone.0001012.
Niu, H., Chen, Y., Ren, Z., Zhang, F., 2008. Sequence analysis of mtDNA D-loop higher-variable region in nine sheep breeds of China. Acta Agriculturae Boreali-occidentalis Sinica 17, 5–9.
Oner, Y., Calvo, J.H., Elmaci, C., 2013. Investigation of the genetic diversity among native Turkish sheep breeds using mtDNA polymorphisms. Tropical Animal Health and Production 45, 947–951.
Pedrosa, S. et al., 2007. Mitochondrial diversity and the origin of Iberian sheep. Genetics Selection Evolution, 39(1): 91.
Ronquist, F., Huelsenbeck, J.P., 2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics, 19(12): 1572–1574.
Saitou, N., Nei, M., 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Molecular biology and evolution, 4(4): 406–425.
Tamura, K., Peterson, D., Peterson, N., Stecher, G., Nei, M., Kumar, S., 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Molecular biology and evolution, 28(10): 2731–2739.
Wang, X., Ma, Y., Chen, H., Guan, W., 2007. Genetic and phylogenetic studies of Chinese native sheep breeds (Ovis aries) based on mtDNA D-loop sequences. Small Ruminant Research, 72(2): 232–236.
Zeng, X.C., Chen, H.Y., Hui, W.Q., Jia, B., Du, Y.C., Tian, Y.Z., 2010. Genetic diversity measures of 8 local sheep breeds in northwest of China for genetic resource conservation. Asian-Australasian Journal of Animal Sciences 23, 1552–1556.
Zhao, Y., Zhao, E., Zhang, N., Duan, C., 2011. Mitochondrial DNA diversity, origin, and phylogenic relationships of three Chinese large-fat-tailed sheep breeds. Tropical Animal Health and Production 43, 1405–1410.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Zhao, E., Yu, Q., Zhang, N. et al. Mitochondrial DNA diversity and the origin of Chinese indigenous sheep. Trop Anim Health Prod 45, 1715–1722 (2013). https://doi.org/10.1007/s11250-013-0420-5
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11250-013-0420-5