Abstract
This study used cultures, polymerase chain reaction (PCR), and immunoperoxidase to examine samples from 216 lungs from sheep and lambs with macroscopic pneumonia lesions for the presence of Mycoplasma species. DNA was extracted from lung tissue samples and broth cultures with the help of a DNA extraction kit and replicated using genus-specific and species-specific primers for mycoplasma. The lung samples were examined by the immunoperoxidase method using hyperimmune Mycoplasma ovipneumoniae serum. The randomly amplified polymorphic DNA (RAPD) test was used for the molecular typing of M. ovipneumoniae isolates. Mycoplasma was isolated in the cultures of 80 (37.03 %) of a total of 216 lung samples. Genus-specific mycoplasma DNA was identified by PCR in 96 (44.44 %) samples in broth cultures and 36 (16.66 %) directly in the lung tissue. Of these 96 cases in which genus-specific identification was made, 57 (59.37 %) were positive for reaction with species-specific primers for M. ovipneumoniae and 31 (32.29 %) for Mycoplasma arginini. The DNA of neither of the latter two species could be identified in the remaining eight samples (8.33 %) where mycoplasma had been identified. As for the immunoperoxidase method, it identified M. ovipneumoniae in 61 of 216 lung samples (28 %). Positive staining was concentrated in the bronchial epithelium cell cytoplasm and cell surface. RAPD analysis resulted in 15 different profiles. Our results suggest that PCR methods could be successfully used in the diagnosis of mycoplasma infections as an alternative to culture method and identifying this agent at the species level.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Pneumonias observed in sheep are of polymicrobial origin, produced by viruses, parasites, chlamydiae and mycoplasmas (Aytuğ 1987; Martin 1983, 1996). Foremost among the agents causing pneumonia among sheep and lambs are Mycoplasma ovipneumoniae, Mannheimia haemolytica, and Pasteurella multocida (Damassa et al. 1992). M. ovipneumoniae causes atypical pneumonia in sheep; it is hypothesized that infection started by this agent sensitizes the animals to infection by M. haemolytica and the Parainfluenza-3 (PI3) virus (Nicholas et al. 2008). These agents jointly cause infection in most cases. Even though Mycoplasma arginini is an important pathogen by itself in sheep and goats, coinfection by M. haemolytica intensifies the pathologic injury of pneumonia (Lin et al. 2008; Nicholas et al. 2008). Mycoplasma species like Mycoplasma mycoides subsp. capri, Mycoplasma capricolum subsp. capricolum and M. capricolum subsp. capripneumoniae, Mycoplasma bovis, and Mycoplasma agalactiae have reportedly been isolated from the respiratory mucosa and lungs of sheep and lambs (Ayling et al. 2004; Erken 2004; Loria et al. 2003; Nicholas 2000).
High morbidity, low mortality, and growth delay generally characterize epidemics of lamb pneumonia. Death can occur due to complications of the infectious condition, like pleuritis, pulmonary abscess, lung consolidation, and acute fibrinous pneumonia. Sheep initially suffer from cough, body temperature increase, loss of appetite, slow growth, and reduced milk production in the ewes. Main clinical signs in sheep and lambs are irregular but persistent, chronic cough, mucopurulent nasal discharge, and severe inflammatory response (Ayling and Nicholas 2007).
Studies in various countries have reported that the most frequently isolated species in sheep and lamb with pneumonia is M. ovipneumoniae, with occasional coinfection by M. haemolytica, PI3 virus, and M. arginini (Lin et al. 2008; McAuliffe et al. 2003a; Nicholas et al. 2008; Parham et al. 2006; Sheehan et al. 2007). M. ovipneumoniae infection is an endemic problem often diagnosed across the world, especially in the USA, Australia, New Zealand, and the UK (Martin 1996). It has been reported that M. ovipneumoniae and M. arginini were isolated from the lungs of healthy sheep and lambs (Alley et al. 1999). Studies performed in Turkey have isolated M. ovipneumoniae, M. arginini, and M. agalactiae in sheep and lamb pneumonia (Erken 2004; Güler 1993).
The microscopic lesions of the sheep lungs infected by M. ovipneumoniae consist of chronic catarrhal bronchitis and bronchiolitis, alveolitis, and atelectasis following bronchial obstruction (Black et al. 1988; Hazıroğlu et al. 1996; Sheehan et al. 2007). Definitive diagnosis requires the isolation and identification of the infectious agent, given the lack of specificity of the clinical and morphological findings. Considering the role of M. ovipneumoniae in the rapid spread of atypical pneumonia in sheep, there is a need for specific but also sensitive and rapid diagnosis of the infected animals. Mycoplasmas are identified by biochemical and serological testing (Erdağ and Türkaslan 1989; Stalheim 1985; Giangaspero et al. 2012). Even though cultures are used for diagnosis in most cases, they are cumbersome and need a long time (Lin et al. 2008; Weiser et al. 2012). The most recent and rapid molecular methods to identify mycoplasmas are 16S rDNA PCR and denaturing gradient gel electrophoresis (McAuliffe et al. 2003b). At the same time, it has been established that mycoplasma antigens can be identified by immunohistochemistry (IH) in lung tissue samples (Hazıroğlu et al. 1996; Sheehan et al. 2007)
Methods like amplified fragment length polymorphism, restriction fragment length polymorphism, random amplified polymorphic DNA (RAPD), and pulsed field gel electrophoresis (PFGE) are used for the molecular typing of M. ovipneumoniae (Harvey et al. 2007; Nicholas et al. 2008; Parham et al. 2006).
The objective of the present study was to identify the mycoplasmas in the lungs of sheep and lambs with pneumonia using cultures, PCR, and immunoperoxidase methods and to perform their molecular typing.
Materials and methods
Sampling
The study material consisted of 180 samples obtained from slaughterhouses in the Elazig Province, in Eastern Turkey, and 36 more that had been brought from different other provinces for diagnosis to the Elazig Veterinary Control and Research Institute. Of these 216 samples, 200 were from sheep and 16 from lambs. The samples were collected in the years 2008–2010, in two runs, most of the samples being from the end of summer or the end of winter.
Infectious agent isolation
Pieces of tissue from the border area between disease lesions and healthy lung were suspended by pounding in a mortar with sand and liquid growth medium. The suspension was inoculated into mycoplasma broth growth media with added Mycoplasma Supplement G (Oxoid, SR0059C). Dilutions of up to 10−5 or the liquid medium was prepared. The inoculated medium was incubated 7–10 days at 37 °C in an incubator with 5 % CO2. The growth medium was checked daily for growth. A loopful of the broth cultures showing mycoplasma growth or color change were inoculated onto Mycoplasma Agar Base media in a 95 % N2 and 5 % CO2 humidified atmosphere at 37 °C and the petri dishes examined in stereomicroscopy at the end of incubation. Colonies suspect for mycoplasma were identified by routine methods (Güler 1993; Yoder 1985).
Primers
The primers used were Mycoplasma genus-specific GPO3 (5′ GGGAGCAAACACGATAGATACCCT 3′) and MGSO (5′ TGCACCATCTGTCACTCTG-TTAACCTC 3′) derived from the 16SrRNA gene (Botes et al. 2005), LMF1 (5′ TGAACGGAATATGTTAGCTT 3′) and LMR1 (5′ GACTTCATCCTGCACTCTGT 3′) species-specific for M. ovipneumoniae (McAuliffe et al. 2003a), and M. arginini species-specific MAGF (5′ GCATGGAATCGCATGATTCCT 3′) and GP4R (5′GGTGTTCTTCCTTATATCTACGC 3′) (Timenetsky et al. 2006).
DNA isolation and PCR
The QIA amp DNA extraction kit (Qiagen) was used for extraction from the lung samples. DNA isolation from mycoplasma broth cultures and organ suspensions was performed according to the users’ instructions for the kit.
A PCR mixture was prepared, containing 5 μL 10× PCR buffer (Tris–HCl 100 mM, pH 9.0, KCl 500 mM, MgCl2 15 mM, 1 % Triton X-100), 250 μM of each of the three deoxynucleotide triphosphates (dNTP), 2 U Taq DNA polymerase (Fermentas, Lithuania), 40 pmol of each of the primers, and 5 μL target DNA in 50 μL. PCR was performed in a Touchdown Thermocycler (Hybaid, UK). DNA amplification for M. ovipneumoniae used, following 5 min preheating at 94 °C, 30 s denaturation at 94 °C, and 30 s hybridization at 55 °C and 72 °C for 30 s, for 30 cycles. The last cycle consisted of 7 min at 72 °C. For M. arginini, the procedure involved 30 s denaturation at 94 °C, 30 s hybridization at 55 °C and 72 °C for 60 s, for 40 cycles; the last cycle was 5 min at 72 °C. DNA amplified by PCR was subjected to agarose gel electrophoresis, performed in a Midi gel electrophoresis tank at 75 V for 1 h, using Tris–boric acid–EDTA as a buffer solution. Following electrophoresis, the gel was stained with ethidium bromide (0.5 μg/mL) for 30 min and the results evaluated on an ultraviolet transilluminator. The band lengths were compared to those of a standard 100 bp DNA ladder (Fermentas). Standard M. ovipneumoniae NCTC 10151 and M. arginini G230 strains were obtained from the Pendik Research Institute Mycoplasma Reference Laboratory.
RAPD test
The 50-mL reaction mix consisted of 5 μL 10× PCR buffer, 1.5 mM MgCl2, 100 μL of each dNTP, 2.5 U Taq DNA polymerase (Fermentas), 1 μM primer, and 5 μL extracted DNA. The sequence of the primers used in RAPD PCR was of the Hum-1 type (5′-3′: TCA CGA TCG A). The thermal conditions were, following an initial denaturation at 94 °C for 5 min, denaturation at 94 °C for 15 s, 60 s annealing at 37 °C, and 90 s synthesis at 72 °C, in a total of 40 cycles. The last cycle consisted of at 72 °C. The PCR-amplified DNA was subjected to agarose gel electrophoresis in an electrophoresis tank at 75 V for 1 h. Following electrophoresis, the gel was stained with ethidium bromide (0.5 μg/mL) for 30 min and the results evaluated on an ultraviolet transilluminator (Parham et al. 2006).
Histologic examination
Lung samples were fixed in 10 % buffered formalin and prepared in the routine way; paraffin blocks were sliced to 4 μm thickness in a microtome and studied in light microscopy following hematoxylin–eosin staining.
Immunohistochemistry study
Paraffin-embedded sections prepared from lung samples were immunochemically stained by an avidin–biotin peroxidase complex (ABC) method. A commercially available kit (Invitrogen) was used for the immunoperoxidase test. Following deparaffinization and rehydration, 4 μM lung sections were kept 10 min in a 3 % hydrogen peroxide solution to block endogen peroxidase activity. The tissues were then kept 5 min in each of two separate phosphate-buffered saline (PBS) solutions. The tissues were treated for 15 min in the microwave oven with a ready-to-use antigen retrieval solution, then cooled at room temperature and kept 10 min in the block solution, and incubated one night at 4 °C in anti-M. ovipneumoniae hyperimmune serum (1/800) (Veterinary Laboratories Agency, UK). The control lung tissues were incubated in distilled water only before washing with PBS. After 10 min treatment with the secondary antibody and washing with PBS, they were incubated 10 min in streptavidin, washed again with PBS, and kept 5 min in 3-amino-9-ethylcarbazole chromogen. After washing with distilled water, the tissues were counterstained with Mayer’s hematoxylin solution and sealed in mounting medium. The anti-M. ovipneumoniae hyperimmune serum was obtained from Dr. Roger Ayling (Veterinary Laboratories Agency, UK)
Results
Culture and PCR results
These results are summarized in Table 1. Mycoplasma was isolated in 80 (37.03 %) of the 216 samples. M. ovipneumoniae was identified in 52 (65 %) of the positive samples and M. arginini in 28 (35 %) of these. PCR with genus-specific primers gave a positive result from the cultures in 96 cases (44.44 %), while PCR performed directly on tissue was positive for the genus in 36 or 16.66 % (see Table 1). Of the 96 cases found positive with genus-specific primers, 57, or 59.37 %, were positive with species-specific primers for M. ovipneumoniae and 31, or 32.29 %, with M. arginini. The bands obtained were 270 bp with the Mycoplasma genus-specific primer (Fig. 1), 361 bp with the specific primer for M. ovipneumoniae, and approximately 545 bp with the species-specific M. arginini primer (Fig. 2). In eight more cases (8.33 %), neither M. ovipneumoniae nor M. arginini DNA could be found in a sample otherwise positive with genus-specific primers. M. ovipneumoniae were found as positive 61 of 216 samples with immunohistochemistry method. Also, nine lung samples which had yielded no growth on culture were found positive by the immunoperoxidase test. All the samples that were positive for M. ovipneumoniae by PCR examination were also positive by IH. At the RAPD analysis of M. ovipneumoniae, 15 different profiles were observed (Fig. 3). The most frequent was profile 1 (eight isolates) and the least frequent profile 7 (two isolates). The prevalence of Mycoplasma species in winter was found higher than the prevalence of summer.
Morphologic findings
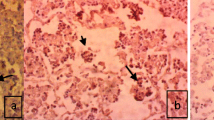
Macroscopic examination showed lesions of chronic bronchopneumonia, consisting of the thickening of the bronchial walls and the presence of consolidation areas on the sectional surfaces of the lungs. These lesions were more pronounced in the cranial lobes, characterized by discoloration reaching from red to brown in these areas. Microscopic examination showed hyperplasia of the peribronchial and peribronchiolar lymphoid tissues, metaplastic and hyperplastic transformation in the bronchial and bronchiolar epithelium, and alveolar septal thickening related to mononuclear cell infiltration (Fig. 4).
Immunohistochemistry findings
The M. ovipneumoniae antigen was specifically shown to be present on the surface or in the cytoplasm of epithelial cells (Fig. 5). The distribution of the staining was in general narrowly related to that of the areas with visible lesions. It was noted that strong positive reactions occurred in lungs of which the agent was isolated, while the immunohistochemical reaction appeared weak in those lungs samples where the agent could not be isolated.
Discussion
M. ovipneumoniae, which is often isolated from sheep with pneumonia, is also found in the respiratory tract of healthy animals (Damassa et al. 1992). It is reported that M. arginini is not a pathogen in its own right but can worsen pneumonias originally due to other microorganisms (Nicholas et al. 2008). In a study of samples from 209 lungs, 126 of which were with pneumonia and 83 healthy; M. ovipneumoniae was isolated in 87 % of the sick and 6 % of the healthy ones in Southern Norway (Bakke 1982). Out of the nasals swabs of 63 sheep with pneumonia, Alley et al. (1975) isolated M. ovipneumoniae in 79 % and M. arginini in 52 %. As for Pasic and Miroslova (1989), they report that mycoplasma was isolated from lung samples of 51 (66.2 %) in 71 sheep with pneumonia; of these, 35 (49.29) were identified as M. ovipneumoniae and 14 (19.71 %) as M. arginini. Sheehan et al. (2007) reporting on 30 lamb lungs with bronchopneumonia have M. ovipneumoniae isolated from 22, M. arginini in four and PI3 virus in nine. Finally, a study by St George and Carmichael (1975) in the USA in 32 nasal swabs from sheep with chronic pneumonia shows M. ovipneumoniae in 18, M. arginini in 24, alongside bacteria like Pasteurella spp., Escherichia coli, and Klebsiella spp.
In Turkey, a study by Güler (1993) conducted in the province of Konya isolated mycoplasmas in 50 (43.47 %) of 115 sheep and goat lungs with pneumonia and 36 of 100 lung samples with pneumonia collected from slaughterhouses; the author reports a higher probability of finding M. arginini than other kinds of microorganisms. Other bacteria, M. arginini foremost, had also been found in this study. Ülgen et al. (1997) report that they isolated Mycoplasma spp. in 7 of 71 sheep lungs with pneumonia collected from the Bursa Provincial Meat and Fish Office slaughterhouse. Baysal and Güler (1992) isolated various microorganisms in 147 out of 186 lamb and calf lungs, 18.46 % of these being Mycoplasma spp. A study by Erken (2004) in the province of Samsun in 200 sheep and lamb lungs with pneumonia yields M. ovipneumoniae in 11 (5.5 %), the same number of M. arginini, and five cases of M. agalactiae, for a total of 27 (13.5 %) cases of mycoplasma isolation.
The ratio of mycoplasma isolation in our study (37 %) is close to that of 36 % reported by Güler (1993). The finding that a majority of cases here were identified as M. ovipneumoniae shows some similarity to studies performed in other countries (Alley et al. 1975; Pasic and Miroslava 1989; Sheehan et al. 2007; Azızı et al. 2011; Besser et al. 2012). These results indicate that the former is more frequent than the latter in our geographic area.
PCR, with its high sensitivity and specificity, presents some advantages over cultures and serologic testing. Timenetsky et al. (2006) studied the presence of mycoplasma in 301 cell cultures by both culture and PCR and found them to be positive in 69 (22.9 %) by culture and 93 (30.9 %) by PCR. They obtained positive cultures in five cases (5.4 %) that were negative by PCR with genus-specific primers. It has been stated that a combination of culture and PCR is the most recommended method (Uphoff and Drexler 2002). The present study showed positivity in 96 (44.44 %) of 216 samples by PCR and 80 (37.03 %) by culture. The DNA of either M. ovipneumoniae or M. arginini could not be identified in eight (8.33 %) cases. The fact that these same eight cases reacted positively to the genus-specific primer shows that species other than these two could be involved. The negative result of cultures in a number of PCR-positive cases suggests either that isolation did not occur because of contamination by other bacteria or that antibiotics could have been used before slaughtering. The PCR test performed directly on organ sample suspensions was positive for genus-specific primers in only 36 (16.66 %) of 216 cases. The low rate of DNA identification in direct determination from the organ samples could conceivably be due to the presence of substances that inhibit PCR.
Many studies of mycoplasma isolation report histological identification of lung lesions and IH recognition of mycoplasma antigens in paraffin sections (Black et al. 1988; Dassanayake et al. 2010; Hazıroğlu et al. 1996; Sheehan et al. 2007). The present study identified typical bronchopneumonia lesions and found specific antigens by the immunoperoxidase technique. The fact that the number of lung samples positive by ABC was larger than that of lungs out of which the microorganism was isolated shows that agents that are no longer vital could be evidenced by immunoperoxidase.
The early isolation and identification of mycoplasmas in an infection is late and cumbersome when compared to that of bacteria. Special media are needed to grow mycoplasmas. Those causing pneumonia in the sheep and lambs have been grown in media like Hayflick’s modified, Friis’ modified, FM4 broth and agar, Eaton’s, SP4, PPLO broth and agar, and Mycoplasma broth and agar (Besser et al. 2008; Güler 1993; Harvey et al. 2007; Lin et al. 2008; McAuliffe et al. 2003a; Sheehan et al. 2007). In this study, M. ovipneumoniae and M. arginini were isolated by culture in Mycoplasma broth and agar supplemented with Mycoplasma Supplement G.
Isolates of M. ovipneumoniae have a high degree of genotypic and phenotypic heterogeneity. A study performed in the UK in 43 isolates of this species from sheep lungs and nasal swabs showed 40 RAPD and the same number of PFGE profiles and it was suggested that there was no relationship at the geographic origins of these isolates (Parham et al. 2006). Investigation by restriction DNA analysis of M. ovipneumoniae isolates in New Zealand sheep with atypical pneumonia showed heterogeneity even within the same herd and it was reported that the presence of multiple strains in the same herd could lead to greater severity of the disease (Ionas et al. 1991; Mew et al. 1985). Harvey et al. (2007) examined 18 strains from 35 sheep by arbitrarily primed PCR and amplified fragment length polymorphism and determined heterogeneity within each herd. In the same study, 57 M. ovipneumoniae isolates showed 15 different profiles by RAPD typing. The present study shows great heterogeneity of M. ovipneumoniae in our geographic area, too.
We conclude that PCR, with its high speed and specificity, can successfully be used as an alternative to cultures that can also make up for negative aspects of bacteriological and serological testing, in diagnosing mycoplasma infections and identifying this agent at the species level. Tissue sections obtained from paraffin blocks with the immunoperoxidase technique showed the presence of mycoplasma antigens, thus confirming the bacteriological results.
References
Alley, M.R., Quinlan, J.R., Clarke, J.K., 1975. The prevalence of Mycoplasma ovipneumoniae and Mycoplasma arginini in the respiratory tract of sheep. New Zealand Veterinary Journal, 23, 137–141.
Alley, M.R., Ionas, G., Clarke, J.K., 1999. Chronic non-progressive pneumonia of sheep in New Zealand—A review of the role of Mycoplasma ovipneumoniae. New Zealand Veterinary Journal, 47, 155–160.
Ayling, R.D., Bashiruddin, S.E., Nicholas, R. A. J., 2004. Mycoplasma species and related organisms Isolated from ruminants in Britain Between 1990 and 2000. Veterinary Record, 155, 413–416.
Ayling, R.D. and Nicholas, R.A.J., 2007. Mycoplasma Respiratory Infections. In: Aitken, I.D. (Ed.), Diseases of Sheep, 4th ed. Blackwell, Oxford. 231–235.
Aytuğ, C.N., 1987. Enzootic pneumonia of the Lambs. Sheep Husbandry and Diseases Symposium, University of Selcuk, Faculty of Veterinary Medicine, 11–12 May, Konya, 113–120.
Azızı, S., Tajbakhsh, E., Rezaıı, A., Nekoueı, S.H., Namjoo, A.R., 2011. The role of Mycoplasma ovipneumoniae and Mycoplasma arginini in pneumonic lungs of slaughtered sheep. Revue Med Vet, 162(6), 310–315.
Bakke, T., 1982. The occurrence of Mycoplasmas and Bacteria in lungs from sheep in Southern Norway. Acta Veterinaria Scandinavica, 23, 235–247.
Baysal, T., Güler, L., 1992. Bacterial agent isolation from kids and lamb with enzootic pneumoniae in Konya Region. Veterinarium, 3, 1–5.
Besser, T.E., Cassirer, E.F., Potter, K.A., Vanderschalie, J., Fischer, A., Knowles, D.P., Herndon, D.R., Rurangirwa, F.R., Weiser G.C., Srikumaran, S., 2008. Association of Mycoplasma ovipneumoniae Infection with Population-Limiting Respiratory Disease in Free-Ranging Rocky Mountain Bighorn Sheep (Ovis canadensis canadensis). Journal of Clinical Microbiology, 46, 423–430.
Besser, T.E., Highland, M.A., Baker, K., Cassirer, E.F., Anderson, N.J., Ramsey, J.M., Mansfield, K., Bruning, D.L., Wolff, P., Smith, J.B., Jenks, J.A., 2012. Causes of Pneumonia Epizootics among Bighorn Sheep, Western United States, 2008–2010. Emerging Infectious Diseases, 18(3), 406–14. doi:10.3201/eid1803.111554.
Black, S.R., Barker, I.K., Mehre, K.G., Crawahaw, G.J., Rosendal, S., Ruhnke, L., Thorsen, J., Carman, S., 1988. An epizootic of Mycoplasma ovipneumoniae Infection in captive Dall’s Sheep (Ovis dalli dalli). Journal of Wildlife Diseases, 24, 627–635.
Botes, A., Peyrot, B.M., Olivier, A.J., Burger, W.P., Bellstedt, D.U., 2005. Identification of three novel Mycoplasma species from ostriches in South Africa. Veterinary Microbiology, 111, 159–169.
Damassa, A.J., Wakenell, P.S., Brooks, D.L., 1992. Mycoplasmas of goats and sheep. The Journal of Veterinary Diagnostic Investigation, 4, 101–113.
Dassanayake, R.P., Shanthalingam, S., Herndon, C.N., Subramaniam, R., Lawrence, P.K., Bavananthasivam, J., Cassier, E.F., Haldorson, G.J., Foreyt, W. J., Rurangirwa, F.R., Knowles, D.P., Beser, T.E., Srikumaran, S., 2010. Mycoplasma ovipneumoniae can predispose bighorn sheep to fatal Mannheimia haemolytica pneumonia. Veterinary Microbiology, 145, 354–359.
Erdağ, O., Türkaslan, J., 1989. General criteria for the diagnosis of Mycoplasma. The Journal of Pendik Veterinary Microbiology, 9, 85–95.
Erken, N., 2004. The isolation and identification of Mycoplasma Species in sheep and lambs that indicates pneumonia in Samsun region (unpublished MSc thesis, Samsun Veterinary Control and Research Institute).
Giangaspero, M., Nicholas, R.A.J., Hlusek, M., Bonfini, B., Osawa, T., Orusa, R., Tatami, S., Takagi, E., Moriya, H., Okura, N., Kato, K., Kimura, A., Harasawa, R., Ayling, R.D., 2012. Seroepidemiological survey of sheep flocks from Northern Japan for Mycoplasma ovipneumoniae and Mycoplasma agalactiae. Trop Anim Health Prod, 44 (3), 395–8.
Güler, L., 1993. The isolation and identification of Mycoplasmas in sheep and goats with pneumonia and determination of antibiotic susceptibility (unpublished PhD thesis, University of Selcuk, Faculty of Veterinary Medicine)
Harvey, M.E., Morrical, D.G., Rosenbusch, R.F., 2007. Sheep flock infections with Mycoplasma ovipneumoniae involve multiple strains. Small Ruminant Research, 73, 287–290.
Hazıroğlu, R., Diker, K.S., Türkarslan, J., Gülbahar, M.Y., 1996. Detection of Mycoplasma ovipneumoniae and Pasteurella haemolytica antigens by an Immunoperoxidase technique in pneumonic ovine lungs. Veterinary Pathology, 33, 74–76.
Ionas, G., Norman, N.G., Clarke, J.K., Marshall, R.B., 1991. Study of the heterogeneity of isolates of Mycoplasma ovipneumoniae from sheep in New Zealand. Veterinary Microbiology, 29, 339–347.
Lin, Y.C., Miles, R.J., Nicholas, R.A.J., Kelly, D.P., Wood, A.P., 2008. Isolation and Immunological detection of Mycoplasma ovipneumoniae in sheep with atypical pneumonia, and lack of A role for Mycoplasma arginini. Research in Veterinary Science, 84, 367–373.
Loria, G.R., Sammartino, C., Nicholas, R.A.J., Ayling, R.D., 2003. In Vitro susceptibilities of field isolates of Mycoplasma agalactiae to Oxytetracycline, Tylosin, Enrofloxacin, Spiramycin and Lincomycin-Spectinomycin. Research in Veterinary Science, 75, 3–7.
Martin, W.B., 1983. Respiratory diseases induced in small ruminants by viruses and Mycoplasma. Rev Sci Tech Off Int Epiz, 2, 311–334.
Martin, W.B., 1996. Respiratory Infections of Sheep. Comparative Immunology, Microbiology and Infectious Diseases, 19, 171–179.
McAuliffe, L., Hatchell, F.M., Ayling, R.D., King, A.I.M., Nicholas, R.A.J., 2003a. Detection of Mycoplasma ovipneumoniae in Pasteurella-Vaccinated sheep flocks with respiratory disease in England. Veterinary Record, 153, 687–688.
McAuliffe, L., Ellis, R.J., Ayling, R.D., Nicholas, R.A.J., 2003b. Differentiation of Mycoplasma species by 16S rDNAPCR and DGGE Fingerprinting. Journal of Clinical Microbiology 41, 4844–4847.
Mew, A.J., Ionas, G., Clarke, J.K., Robinson, A.J., Marshall, R.B., 1985. Comparison of Mycoplasma ovipneumoniae Isolates using bacterial restriction endonuclease DNA analysis and SDS-PAGE. Veterinary Microbiology, 10, 541–548.
Nicholas, R.A.J., 2000. Improvement in the diagnosis and control of diseases of small ruminants caused by Mycoplasmas. Small Ruminant Research, 45, 145–149.
Nicholas, R.A.J., Ayling, R.D., Loria, G.R., 2008. Ovine Mycoplasmal infections. Small Ruminant Research, 76, 92–98.
Parham, K., Churchward Colin, P., Mcauliffe, L., Nicholas, R.A.J., Ayling, R.D., 2006. High level of strain variation within the Mycoplasma ovipneumoniae population of the UK has Implications for disease diagnosis and management. Veterinary Microbiology, 118, 83–90.
Pasic, S., Miroslava, P., 1989. Proucavanje Ovinih Mikoplazmi u Bosni i Hercegovini. Veterinaria, 38, 307–311.
Sheehan, M., Casidy, J.P., Brady, J., Ball, H., Doherty, M.L., Quinn, P. J., Nicholas, R.A.J., Markey, B.K., 2007. An aetiopathological study of chronic bronchopneumonia in lambs in Ireland. The Veterinary Journal, 173, 630–637.
Stalheim, O.H.V., 1985, Mycoplasmas of animals. In: G. R. Carter (ed) Diagnostic Procedures in Veterinary Bacteriology and Mycology. Iowa State University Press, Iowa. 263–288.
St George, T.D., Carmichael, L.E., 1975. Mycoplasma ovipneumoniae from sheep with chronic pneumonia. Veterinary Record, 97, 205–206.
Timenetsky, J., Santos, L.M., Buzinhani, M., Mettifogo, E., 2006. Detection of multiple Mycoplasma infection in cell cultures by PCR. Brazilian Journal of Medical and Biological Research, 39, 907–914.
Uphoff, C.C., Drexler, H.G., 2002. Comparative antibiotic eradication of Mycoplasma infections from continuous cell Lines. In Vitro Cellular & Developmental Biology—Animal, 38, 86–89.
Ülgen, M., Sönmez, G., Aydın, F., 1997. The examination microbiological and histopathological of pneumonia in lamb. Journal of Etlik Veterinary Microbiology, 9, 55–70.
Weiser, G.C., Drew, M.L., Cassirer, E.F., Ward, A.C., 2012. Detection of Mycoplasma ovipneumoniae in bighorn sheep using enrichment culture coupled with genus- and species-specific polymerase chain reaction. J Wildl Dis, 48(2), 449–53. doi:10.3201/eid1803.111554.
Yoder, H.W., 1985. Laboratory methods for Mycoplasmas, diagnostic procedures in Veterinary Bacteriology and Mycology. Ed. G.R. Carter. Iowa State University Press. Iowa. 489–495.
Acknowledgments
This investigation was supported by the General Directorate of Agricultural Research of the Ministry of Agriculture and Village Affairs of the Republic of Turkey (project number TAGEM/HS/08/02/08/134).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution License which permits any use, distribution, and reproduction in any medium, provided the original author(s) and the source are credited.
About this article
Cite this article
Kılıc, A., Kalender, H., Eroksuz, H. et al. Identification by culture, PCR, and immunohistochemistry of mycoplasmas and their molecular typing in sheep and lamb lungs with pneumonia in Eastern Turkey. Trop Anim Health Prod 45, 1525–1531 (2013). https://doi.org/10.1007/s11250-013-0394-3
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11250-013-0394-3