Abstract
Agricultural crops are susceptible to many diseases caused by various pathogens, such as viruses, bacteria and fungi. This paper reviews the general principles of plant protection against pathogens, as well as the role of iron and antimicrobial peptide metabolism in plant immunity. The article highlights the principles of antibacterial, fungicidal and antiviral action of lactoferrin, a mammalian secretory glycoprotein, and lactoferrin peptides, and their role in protecting plants from phytopathogens. This review offers a comprehensive analysis and shows potential prospects of using the lactoferrin gene to enhance plant resistance to various phytopathogens, as well as the advantages of this biotechnological approach over existing methods of protecting plants against various diseases.
Similar content being viewed by others
Introduction
Growth of the global population triggers a significant rise in demand for agricultural products and cultivation of crops. However, since prehistoric times until nowadays, mankind has been suffering from large-scale yield losses (Cerda et al. 2017; Savary et al. 2012; Teng et al. 1984; Chaloner et al. 2021; Peng et al. 2021). Plant crops are susceptible to numerous diseases caused by various pathogens from viruses and viroids to bacteria, fungi and protozoa. Plant diseases can cause both mild symptoms and large-scale yield losses; furthermore, due to the accumulation of toxins, phytopathogenic microorganisms shorten the shelf life of food products and deteriorate their nutritional value (Shuping et al. 2017). The spread of phytopathogens is difficult to control due to their ability to rapidly mutate and the emergence of new highly virulent strains. The emergence of highly variable genotypes of phytopathogens is facilitated by a high planting density and genetic homogeneity of crops grown in large areas. Low selective pressure promotes the growth of genetic variability of the pathogen, which can lead to infecting of the same plant with different genotypes of a certain pathogen and increase the virulence of microorganisms competing for a limited source. In addition, the globalization of the world economy and climate change are contributing to the spread of pathogens to new territories (McDonald et al. 2016).
To counteract the challenge, classical methods of crop protection, such as crop rotation or chemical protection with pesticides could be applied. Besides, plant breeding methods allow to create the plant cultivars resistant to several diseases, as well as the post-harvest protection could also be applied. However, the classical methods of plant protection are usually inefficient due to the high genetic variability of pathogens (McDonald et al. 2016; Garfinkel et al. 2019). The intensive development of biotechnology over the last three decades opens up promising opportunities for increasing the resistance of agricultural plants to phytopathogens. Plant biotechnology offers a powerful strategy which may ensure the availability and high quality of food for the whole twenty-first century. Genetic engineering of plants, including gene editing as an alternative to breeding and the use of pesticides, could provide a comprehensive long term protection of crops against bacterial, fungal and viral phytopathogens. The transfer of genes encoding antimicrobial proteins into plant genomes is a promising means of protecting plants from various diseases. Such genetic engineering approach is considered potentially safe for the environment and human health (Bruce et al. 2012; Ceasar and Ignacimuthu 2012; Grant et al. 2013; Owen et al. 2010; Razzaq et al. 2021; Varshney et al. 2021).
Lactoferrin is a well-studied mammalian iron-binding protein with strong antimicrobial and immunomodulatory activity and low allergenicity. Due to the diverse beneficial properties of this protein, the expression and production of recombinant lactoferrin in the genomes of agricultural plants can be useful for various pharmaceutical applications, biological enrichment of products (Adleranova et al. 2008; Lakshman et al. 2013; Stefanova et al. 2008; Tanasienko et al. 2011; Yemets et al. 2014), as well as for protection of plants from various pathogens (Buziashvili et al. 2020a, 2020b; Chahardoli et al. 2018; Fukuta et al. 2012; Han et al. 2012; Lee et al. 2002; Malnoy et al. 2003; Mitra and Zhang 1994; Nguyen et al. 2011; Stefanova et al. 2013a, b; Takase et al. 2005; Zhang et al. 1998). This article discusses the general principles of plant protection strategies against pathogens, as well as the role of iron and antimicrobial peptide (AMP) metabolism in plant immunity. Examples of successful application of the lactoferrin gene to transform different plant species and increase their resistance to diseases, as well as the challenges and opportunities associated with the use of lactoferrin for crop protection, are also discussed.
The plant immune response and plant defense mechanisms against diseases
Most plants require solid (soil) and gaseous (atmosphere) environments simultaneously, in which they are exposed to various biotic and abiotic factors. The average density of bacterial cells in the soil is 108/g−1, apart from fungi, nematodes and other pathogens. Thus, a strong immune system is required to protect plants against various biotic and abiotic stresses (Raynaud et al. 2014). Unlike animals, plants lack mobile immune cells and adaptive immunity. Instead, they rely on the innate immune system (Han 2019; Jones and Dangl 2006; Zhang et al. 2020).
In general, the plant immune response can be characterized by an accurate ‘zig-zag’ scheme proposed by Jones and Dangl (2006). According to this scheme, the first stage of an immune response begins with the recognition of conserved macromolecular complexes named Pathogen-Associated Molecular Patterns (PAMPs) by transmembrane Pattern Recognition Receptors (PRRs), which also recognize the molecules emanating from the sites of damage (Damage-Associated Molecular Patterns (DAMPs)) (Andersen et al. 2018; Henry et al. 2013; Zhang et al. 2020). Receptors transmit the signal through molecular cascades leading to the expression of Pathogenesis-Related (PR) genes. This reaction, called PAMP-Triggered Immunity (PTI), may be sufficient to stop the colonization of plant cells by pathogens. In stage 2, called Effector-Triggered Susceptibility (ETS), the pathogen produces effectors that interfere with the PR genes and can spread around the site of infection and feed on its tissues making the plant susceptible to a disease. In stage 3, the effector molecules of the pathogen are recognized by internal receptors of the host, such as receptors with Nucleotide-Binding domains and Leucine-Rich Repeats (NB-LRR), which leads to Effector-Triggered Immunity (ETI). Receptors that recognize pathogen effectors are encoded by R(resistance)-genes. The Effector-Triggered Immunity is an enhanced version of the PTI response that often results in a hypersensitivity response and a programmed cell death in infected tissues. As in stage 1, this reaction may be sufficient to provide resistance. In stage 4, the pathogen avoids ETI, surpassing the receptors for pathogen recognition or acquiring additional effectors, which leads to the emergence of new R-alleles (Jones and Dangl 2006).
The mechanisms of Effector-Triggered Immunity can be more meticulously explained by the gene-for-gene model which describes the interaction between the single dominant avirulence genes of the pathogen (Avr) and plant R-genes (Rao et al. 2021). Recognition of pathogen effectors by R-proteins leads to an incompatible interaction and disease resistance. If such recognition doesn’t occur (e.g., due to the lack of appropriate R-genes), the interaction becomes compatible, the pathogen is virulent, and the plant is called susceptible. R-gene expression as a result of ETI often leads to a Hypersensitive Response (HR) (Rao et al. 2021).
The plant protection mechanisms may vary depending on the pathogen infection strategy. According to the infection strategy, plant pathogens can be identified as biotrophs, hemibiotrophs and necrotrophs. Pathogens of the first group (biotrophs) feed and reproduce themselves on living tissues; necrotrophs kill the host and consume organic matter; in the early stages of infection, hemibiotrophs feed on living tissues, but later the host dies. It is important to note that most bacterial plant pathogens such as Ralstonia solanacearum, Clavibacter michiganensis, Pseudomonas syringae, Erwinia amylovora, are commonly referred to as biotrophs or hemibiotrophs (Jacobs et al. 2013; Mas Muniroh 2018; Kraepiel and Barny 2016; Emeriewen et al. 2019), whereas fungal pathogens, e.g. Fusarium sp., Rhizoctonia solani, Botrytis cinerea, Phytophthora infestans, etc. are known as necrotrophs or hemibiotrophs (Abdelghany et al. 2022; Boddy 2016; Rauwane et al. 2020; Nowicki et al. 2011). Plant immune responses differ for each of these groups (Glazebrook et al. 2005). Plants infected by biotrophs or hemibiotrophs develop a Hypersensitive Response (HR) which leads to the production of Reactive Oxygen Species (ROS), oxidative burst and apoptosis. These reactions usually inhibit the reproduction of biotrophic and hemibiotrophic pathogens and lead to Systemic Acquired Resistance (SAR). The hypersensitive response is mediated by salicylic acid (SA) (Conrath 2006; Durrant et al. 2004; Vlot et al. 2020). In contrast, the protective strategies against necrotrophs include cell wall modification, wax synthesis, and production of antimicrobial compounds such as chitinases, protease inhibitors, phytoalexins and various antimicrobial peptides (AMPs). These reactions are regulated by jasmonate (JA) and ethylene (ET) (Glazebrook et al. 2005). Moreover, an alternative mechanism of protection against necrotrophs, the Induced Systemic Resistance (ISR), is also activated by JA and ET (de Vleesschauwer et al. 2009; Vallad et al. 2004; Vlot et al. 2020). This reaction is driven by mutualistic species of Plant Growth-Promoting Rhizobacteria (PGPR), such as Pseudomonas, Serratia and Bacillus genera, or by Plant Growth-Promoting Fungi (PGPF) (e.g., non-pathogenic strains of Fusarium sp.). These microorganisms produce siderophores, iron-binding molecules, that promote competition between symbiotic and pathogenic microorganisms for iron ions in conditions of iron-deficiency, and as a result, ISR enhances the systemic resistance of plants to a wide range of phytopathogens (Vallad et al. 2004; van Loon et al. 2005; Choudhary et al. 2007; Pieterse et al. 2014; Romera et al. 2019). Thus, iron metabolism is essential for plant immunity.
Another method of disease defense is mediated by antimicrobial peptides (AMPs). AMPs, usually small molecules of 12–50 amino acids, are widely present in all living organisms (Jung et al. 2014; López-García et al. 2012). The first antimicrobial peptide, gramicidin from a soil Bacillus sp. strain, was described by Hotchkiss and Dubos in 1940. Among the first animal antimicrobial peptides described were defensin and bombinin from the rabbit leucocytes and epithelial cells, as well as lactoferrin from cow’s milk (see review Jung et al. 2014). In plants, they are termed PR proteins. Plant PR proteins are small peptides of about 5–75 kDa, but can also include enzymes or larger proteins. They are grouped into 17 families depending on their activities. Some examples of the PR peptides and proteins are chitinases, glucanases, Thaumatin-Like Proteins (TLPs), proteinase inhibitors, peroxidases, Ribonuclease-Like Proteins (RLPs), defensins, thionins, Lipid Transfer Proteins (LTPs) and Oxalate Oxidases (OXOs) (López-García et al. 2012; Moosa et al. 2017).
All AMPs expressed in different organisms have common features such as basic, amphipathic and cysteine-rich peptides with rigid tertiary conformation stabilized by disulfide bonds. Depending on their structure, the AMPs could be classified into 4 groups: (1) α- helical peptides, (2) ß-sheet peptides with two or more disulfide bonds, (3) Peptide molecules with a single hairpin or loop containing disulfide bonds and (4) Long peptides (Jung et al. 2014; Li et al. 2021). All AMPs specifically target bacterial membranes which differ from the eukaryotic ones by negatively charged extracellular phospholipids (Jung et al. 2014; López-García et al. 2012). The mechanism of action of AMPs depends on their structure. Most α-helix peptides disrupt bacterial cells forming carpet-like clusters or bundles of barrel-shaped pores. The β-sheet peptides usually insert into the lipid bilayer and form toroidal pores (Jung et al. 2014; Li et al. 2021). Some of the AMPs, such as plant defensins, just like antibiotics, inhibit enzyme activity and protein synthesis in pathogen cells (Jung et al. 2014; Li et al. 2021).
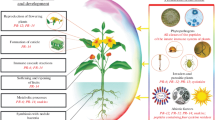
Since some phytopathogens can evade the gene-for-gene protection mechanisms, an effective strategy to enhance the resistance of plants to diseases is to transform plants with genes encoding AMPs or non-plant proteins with antimicrobial activity against a wide range of pathogens (Marcos et al. 2008; Rommens and Kishore 2000; Sinha and Shukla 2019). The transformation of plants with AMPs or antimicrobial proteins of various origin can enhance the ETI of plants not only to necrotrophic and hemibiotrophic, but also to biotrophic phytopathogens through the ISR mechanism. Examples of AMPs used for the genetic transformation of plants are α-helix cecropin from giant silk moths, magainin from tropical amphibians, β-sheet defensin or protegrin, linear melittin from bee venom, and more. However, some AMPs and antimicrobial proteins can be toxic to potential consumers, such as cecropin, which has hemolytic activity (Osusky et al. 2000; Marcos et al. 2008). In addition, antimicrobial proteins of various origins, such as chitinase, glucanase, defensin, thionine, lysozyme (Patil et al. 2012; Jung and Kang 2014; Ceasar and Ignacimuthu 2012; Moosa et al. 2017) can be used to transform plants to protect against certain diseases. Another promising category of genes for strengthening resistance of agricultural plants are the genes of lactoferrin and lactoferrin-derived peptides (Ali et al. 2018; Yemets et al. 2014). Many useful properties of lactoferrin and lactoferrin-derived AMPs are described in detail in the next section. A schematic representation of plant protection strategies and the putative role of lactoferrin in the plant immune response is shown in Fig. 1.
Schematic representation of the key events in plant immune response (indicated as PTI, ETS, ETI, SAR, ISR), gene groups involved in defense strategies (green boxes), the cellular outcomes (blue boxes) and the functions of lactoferrin (red box) which enhance plant’s resistance to phytopathogens (underlined). PAMPs—Pathogen-Associated Molecular Patterns; PRR—Pattern Recognition Receptors; PTI—PAMP-Triggered Immunity; ETS—Effector-Triggered Susceptibility; R-genes—Resistance genes; NB-LRR receprots—receptors with nucleotide-binding domains and leucine-rich repeats; PR-genes—Pathogenesis-Related genes; SA—salicylic acid; JA—jasmonate; ET—ethylene; HR—Hypersensitive Response; ROS—reactive oxygen species; SAR—Systemic Acquired Resistance; ETI—Effector-Triggered Immunity; ISR—Induced Systemic Resistance; PGPB—Plant Growth-Promoting Bacteria
Structure and antimicrobial activity of lactoferrin and lactoferrin peptides
Over the last 20 years, many beneficial properties of milk proteins and protein hydrolysates have been discovered, such as antimicrobial, antifungal, antithrombotic, immunomodulatory, antiviral and anti-inflammatory activities (Adleranova et al. 2008; Giansanti et al. 2016; Kell et al. 2020; Wang et al. 2020a; Oda et al. 2021; Zhang et al. 2021). Physical and chemical properties and bioactivities of milk proteins and peptides, such as lactalbumin, lactoglobulin and lactoferrin, are listed in the MilkAMP database (Bruni et al. 2016).
Lactoferrin is a mammalian iron-binding glycoprotein with a strong antimicrobial activity which plays a significant role in innate immunity. It is a basic protein with an isoelectric point at pI = 8.7. The polypeptide chain of lactoferrin consists of approximately 700 amino acid residues (711 for human lactoferrin and 689 for bovine lactoferrin) folded into two homologous subunits connected with a flexible α-helix, each subunit containing 2 domains (N1, C1, N2, C2) (Adleranova et al. 2008; Giansanti et al. 2016). There is one iron-binding site in each subunit where Fe 3+ and Fe 2+ ions are bound in the presence of two carbonate (CO32−) ions. The protonation of the interdomain H-bonds leads to dissociation of the Fe-lactoferrin complex. Unlike transferrin, lactoferrin is able to keep the ferric ions bound at pH below 4.5, and the affinity of lactoferrin to Fe at low pH is 300 times stronger than that of transferrin (Abdallah et al. 2000; Mantel et al. 1994). This feature is important in case of inflammation sites where lactoferrin firmly binds iron and thus prevents its absorption by the pathogenic bacteria (Adleranova et al. 2008). The molecular weight of lactoferrin varies from 76 to 80 kDa. This difference in molecular weight depends on the state of glycosylation of the protein. Lactoferrin has three glycosylation sites at Asn138, Asn479 and Asn624, respectively. The two first sites of N-glycosylation usually carry complex N-glycans, while the third one (Asn624) is mostly non-glycosylated (Spik et al. 1982). Lactoferrin is capable of binding not only Fe3+ but also other metal ions, such as Ca2+, Mg2+, Cu2+, Mn3+, Zn2+, etc. (Adleranova et al. 2008; Giansanti et al. 2016), although its affinity to ions of other metals is much lower than for Fe3+. Interaction with other ions impacts the structure, function and conformation of lactoferrin. For example, the affinity of lactoferrin to Fe3+ increases in the presence of Zn2+ and Cu2+ (Hakkansson et al. 1995); in the presence of 10 mM of CaCl2 lactoferrin may form tetramers (Bennett et al. 1981). Besides the metal ions, lactoferrin can bind to various organic molecules such as lipopolysaccharides, heparin and glycosaminoglycans (Adleranova et al. 2008; Giansanti et al. 2016).
Nowadays, much work is devoted to investigation of antimicrobial activity of lactoferrin and lactoferrin peptides. Since not only the whole lactoferrin molecule but also its short fragments reveal antimicrobial activity, the conclusion can be made that the antimicrobial properties of lactoferrin depend both on its iron-binding ability and on the structure of peptides derived from lactoferrin. Within a wide range of peptides isolated from lactoferrin, the antimicrobial activity has been confirmed only for three of them: Lf (1–11), lactoferricin and lactoferrampin. All these peptides are located at the N-terminus of the Lf molecule and reveal a stronger antimicrobial effect than intact Lf (Bruni et al. 2016; Bielecka et al. 2022; Giansanti et al. 2016; Gruden et al. 2021; Sinha et al 2013).
Lf (1–11) is a basic peptide which contains the sequence from the first to the eleventh N-terminal amino acids of lactoferrin. Human Lf (1–11) (GRRRSVQWCAV) contains a highly variable loop and a short β-sheet and is rich in arginine so it can be classified as AMP of the 3rd Group. Antimicrobial activity of Lf (1–11) is due to the strong cationic effect of the peptide and the presence of hydrophobic valine- (V6) and tryptophan- (W8) residues in its structure. These features ensure the disruption of the cellular and mitochondrial membranes of bacterial or fungal pathogens by Lf (1–11). (Bruni et al. 2016; Bielecka et al. 2022; Giansanti et al. 2016; Gruden et al. 2021; Sinha et al. 2013).
Lactoferrampine (Lfampin) is another Lf peptide consisting of 16 amino acid residues (268–284) of the N1 domain of lactoferrin. Similarly to Lf (1–11), it has a hydrophobic domain and is positively charged. Lfampin belongs to the AMPs of the 1st Group as it has an α-helical motif which plays a key role in the membrane-associated activities of Lf (Bruni et al. 2016; Bielecka et al. 2022; Giansanti et al. 2016; Gruden et al. 2021; Sinha et al 2013).
Human lactoferricin (Lfcin) is the longest biologically active Lf peptide consisting of 47 amino acids (1–47). It is formed by pepsin-mediated cleavage of lactoferrin. Both Lfcin and Lfampin are closely located in the tertiary structure of the Lf molecule. Bovine Lfcin is shorter and located more distally (17–41) but has stronger bactericidal properties compared to human Lfcin. Both Lfcins belong to the 3rd Group of AMPs as they are positively charged; also, both Lfcins contain a loop of 18 amino acid residues as well as a stack of hydrophobic residues. The mechanism of antibacterial activity of Lfcin is similar to that of Lfampin and is associated with the disruption of cellular membranes of the microorganisms (Bruni et al. 2016; Bielecka et al. 2022; Giansanti et al. 2016; Gruden et al. 2021; Sinha et al. 2013).
Antibacterial activity of lactoferrin and lactoferrin peptides has been confirmed against a wide range of bacterial pathogens in humans, animals and plants (Bruni et al. 2016; Bielecka et al. 2022; Gruden et al. 2021; Jahani et al. 2015; Lakshman et al. 2013; Stefanova et al. 2008; Yemets et al. 2014; Kell et al. 2020; Zhang et al. 2021). Moreover, the fungicidal activity of Lf and Lf peptides against phytopathogenic fungi has been carefully investigated. According to Munoz et al. (2006), the fungistatic and bactericidal activities of bovine Lfcins (17–31 and 20–25) both in vitro and in vivo have been established against a wide range of plant fungal pathogens, such as Fusarium oxysporum which causes tomato fusarium wilt; Botrytis cinerea, the causative agent of grey mold; rice fungus Magnaporthe grisea; post-harvest pathogens Penicillium italicum and P. digitatum known to infect citrus fruits, and apple pathogen Penicillium expansum; Alternaria sp. as a fungus resistant to commonly used fungicides, and the model filamentous fungus Aspergillus nidulans. The antifungal effects of lactoferricins varied from inhibition of the mycelium growth to permeation of cell walls and plasma membranes of hyphae but not conidia. In another study (Lahoz et al. 2008), the fungistatic activity of iron-free bovine lactoferrin (bLf) was studied against 11 phytopathogenic fungal species: Aspergillus niger, Alternaria alternata, Colletotrichum lindemuthianum, Fusarium solani, Gliocladium roseum, Penicillium expansum, Phoma exigua, Rhizoctonia solani, Sclerotinia sclerotiorum, Sclerotium rolfsii and Trichoderma viride. As a result, inhibition of the mycelium growth in vitro was detected for most of the studied species, except for A. alternata, G. roseum, F. solani and C. lindemuthianum. The authors presumed that these four fungal species were insensitive to lactoferrin either because their siderophores might have stronger affinity to Fe than those of bLf, or because the concentration of Fe in the medium was sufficient for their growth. In addition, the antiviral activity of lactoferrin was shown against a wide range of human viral infections (Berlutti et al. 2011; Wakabayashi et al. 2014), including COVID-19 (see reviews Wang et al. 2020b; Kell et al. 2020), and against some plant viruses such as Tomato Yellow Leaf Curl Virus (TYLCV) (Abdelbacki et al. 2010). In summary, the antiviral effect of lactoferrin is carried out by binding either to heparin sulphate glycosaminoglycan viral receptors or to the viral particles, which prevents the latter from penetration into the host cells. The mechanism of antibacterial and fungistatic activity of Lf and its peptide derivatives is primarily to destabilize cell membranes, with iron sequestration playing a secondary role (Fernandes et al. 2017).
Transformation of different plant species with the full-length human lactoferrin gene
The biotechnological transfer of a lactoferrin gene into plant genomes and its expression can enhance the natural immune protection of plants by sequestering iron and directly destroying pathogen cells. Therefore, here we highlight and review the available data on the genetic transformation of plants with different lactoferrin genes and the production of plant lines with increased resistance to certain phytopathogens (Table 1). The resistance of plants to phytopathogens has been thoroughly studied in (Mitra and Zhang 1994; Zhang et al. 1998; Lee et al. 2002; Malnoy et al. 2003; Takase et al. 2005; Nguyen et al. 2011; Fukuta et al. 2012; Han et al. 2012; Stefanova et al. 2013a, b; Chahardoli et al. 2018; Buziashvili et al. 2020a,b) and is comprehensively discussed in this paper. We also focus our attention on the advantages and disadvantages of using either full-length lactoferrin genes or the genes of lactoferrin peptides, on the type of the promoter controlled gene expression as well as the origin (human or bovine) of the lactoferrin gene to increase plant resistance to various pathogens.
The first report on transformation of tobacco with a human lactoferrin gene was published by Mitra and Zhang (1994). In this work, tobacco callus was transformed with the hLf gene, which was expressed under control of the CaMV35S promoter. The antibacterial activity of total protein extracts from transgenic callus was confirmed against 4 different phytopathogenic bacteria: Xanthomonas campestris pv. phaseoli, Pseudomonas syringae pv. phaseoli, Pseudomonas syringae pv. phaseolicola and Ralstonia solanacearum. In 1998, Zhang et al. published another study on obtaining whole tobacco plants expressing hLf. This study demonstrated for the first time the ability of lactoferrin to enhance the resistance of transgenic plants to phytopathogenic bacteria (Ralstonia solanacearum). As a result, the transgenic lines delayed the development of withering symptoms by 6–32 days while the control lines totally wilted by the 11th–12th day post-inoculation. The estimated content of full-length and truncated hLf in transgenic lines was 0.1 to 0.9% of the total soluble protein (TSP). Therefore, such fluctuations in the delay of bacterial wilting symptoms between the transgenic lines could be due to the differences in lactoferrin content.
Transgenic plants of alfalfa (Medicago sativa) expressing human lactoferrin were obtained by Stefanova et al. (2013a). For this purpose, the highly embryogenic Bulgarian cultivar Obnova-10 was transformed with the hLf gene controlled by the CaMV35S promoter. The content of human lactoferrin ranged from 0.0035 to 0.0047% of TSP. The authors explained that the low level of lactoferrin expression, as well as the variations in morphology and growth intensity, were caused by stressful conditions during the cultivation which affected plant growth, development, and protein synthesis. The transgenic plants were tested for resistance to Pseudomonas syringae pv. syringae, that causes bacterial canker, and Clavibacter michiganensis, that causes wilting. The transgenic plants revealed a higher resistance and barely pronounced symptoms compared to controls (Stefanova et al. 2013a), which could be attributed to the expression of hLf. In further studies, the authors investigated the effect of hLf expression on leaf cell morphology (Stefanova et al. 2013b). In particular, the hLf-expressing plants had enlarged pavement cells and a lower stomatal density than control plants. Besides that, the transgenic plants had smaller leaves and lower chlorophyll content. The authors supposed that the observed morphological changes resulted from the iron chelation and inhibition of the cell activity by recombinant lactoferrin. The authors also supposed that the reduction of the number of stomata led to a lower number of infection sites for the pathogens, which could be one of the mechanisms of strengthening the resistance to diseases in transgenic plants (Stefanova et al. 2013b).
One of the most economically detrimental bacterial pathogens with a wide range of hosts is Ralstonia solanacearum causing bacterial wilt of tomatoes. To improve the resistance to R. solanacearum, Lee et al (2002) carried out the Agrobacterium-mediated transformation of tomato line F7926-96, susceptible to bacterial wilt, with the hLf gene driven by the CaMV35S promoter. The results indicated that within 56 days of the experiment, 44–55% of transgenic plants did not show any symptoms of bacterial wilt until the ripening of the fruits, while the control plants completely withered by the 26th day post-inoculation (dpi). However, the delay in wilting varied between individual transgenic tomato plants and lines, as in the similar work on tobacco by (Zhang et al. 1998), but the level of lactoferrin expression in transgenic tomato lines was not indicated in this study. Nevertheless, this work shows the potential of using the hLf gene to increase the resistance of transgenic tomato lines to such a highly virulent bacterial pathogen as R. solanacearum (Lee et al. 2002).
Recently, Buziashvili et al. (2020a, b) studied the transformation of tomato and potato lines with the hLf gene driven by the CaMV35S promoter in order to enhance their resistance to highly virulent bacterial and fungal phytopathogens. In the first study, tomato cultivars Money Maker and Lahidny were used, which are susceptible to bacterial and fungal diseases (Buziashvili et al. 2020b). The content of hLf in transgenic plants was estimated at approximately 0.02 and 0.04% of TSP (4.8 and 8.3 µg/g of fresh weight) for cvs. Money Maker and Lahidny, respectively. In another work, Buziashvili et al. (2020a) reported a transfer of the hLf gene into four Ukrainian potato cultivars, Vernisage, Svitanok Kyivskyi, Levada and Zarevo. It was found that the content of lactoferrin in transgenic potato lines was higher than in tomato lines (about 0.05% of TSP). It was shown that the samples prepared from the stems and leaves of transgenic tomato and potato lines inhibited the growth of bacterial pathogens R. solanacearum, as well as Clavibacter michiganensis subsp. michiganensis causing bacterial canker of tomato, and C. michiganensis subsp. sepedonicus causing brown rot of potato. Moreover, the results of the in vitro inoculation assays showed the enhancement of the resistance of transgenic tomato and potato lines to P. infestans on average from 1 to 7 points of the 9-point scale compared to control (Buziashvili et al. 2020a, b). The results of these studies (Buziashvili et al. 2020a, b) indicate that the expression of human lactoferrin in both transgenic tomato and potato plants could increase their resistance not only to bacterial pathogens such as R. solanacearum and C. michiganensis, but also to highly aggressive fungal pathogens such as P. infestans.
Takase et al. (2005) investigated the resistance to bacterial, fungal and viral pathogens of the rice plants cv. Nipponbare transformed with the genes of N-lobe (hLfN) and full-length hLf under the control of the CaMV35S promoter. The content of lactoferrin in leaves was estimated at 0.1 mg/g, and the hLf expression reduced the growth of transgenic plants by 10–20% compared to the control. In addition, the authors examined the resistance of transgenic plants expressing both truncated and full-length lactoferrin to bacterial (Pseudomonas plantarii), fungal (Pyricularia orysae) and viral (Rice dwarf virus) pathogens. Infection with P. plantarii did not affect plants at the germination stage, but after germination, the transgenic plants expressing either hLf or hLfN showed higher resistance to bacterial seedling blast than non-transgenic plants. Test on resistance to a Rice dwarf virus showed that in 20 days both transgenic and control plants developed symptoms of virus infection. However, the transgenic lines expressing hLf, but not an hLfN gene, developed symptoms 2–3 days later than the control ones. The study of the resistance to fungal pathogen P. oryzae showed no difference in susceptibility to the fungus: both transgenic and control plants developed severe lesions. Nevertheless, the transgenic rice lines expressing hLf were characterized by an increased resistance to bacterial and viral pathogens. These results show a positive aspect of hLf expression in rice making it resistant to disease and non-carrying phytopathogenic toxins, which means a better quality of rice grains containing Lf (Takase et al. 2005).
This suggests that transformation of plants with the full-length hLf gene and its expression in the plant genome could change the morphology of transgenic plants, as it was indicated in the works of Stefanova et al. (2013a) and Takase et al. (2005). Moreover, the studies of Zhang et al. (1998) and Lee et al. (2002) reported variability in the resistance of transgenic tobacco and tomato plants to R. solanacearum, and the work of Takase et al. (2005), demonstrated the absence of resistance to the fungus Pyricularia orysae. However, in the works of Zhang et al. (1998), Lee et al. (2002), Buziashvili et al. (2020a, b), Stefanova et al. (2013a) and Takase et al. (2005), the plants, transformed with the hLf gene, showed enhanced resistance to highly virulent bacterial (R. solanacearum; C. michiganensis; P. syringae; P. plantarii), fungal (P. infestans) and viral (Rice dwarf virus) pathogens (Table 1).
Transformation of plants with human lactoferrin-derived genes
The effect of the hLfN gene expression in Nicotiana bentamiana plants on their post-infection resistance to viral pathogens was studied by (Li et al. 2004). In this study, agroinfiltration of transgenic tobacco plants carrying the sequence of the N-lobe of the hLf gene with Potato virus X (PVX) was carried out. The results showed that both control and transgenic tobacco plants expressing the hLfN gene, agroinfiltrated with PVX, developed symptoms of Potato virus X disease. This means that the N lobe of the hLf gene did not ensure post-infection resistance to plant viruses. By the way, the content of produced hLfN was 0.6% of TSP, which was higher compared to the previous studies (Salmon et al. 1998; Mitra and Zhang 1994). Total protein extracts obtained from the plants with the hLfN were almost 2 times more effective in inhibiting the growth of E. coli, than S. aureus and S. typhimurium, while no bacteriostatic effects were observed for the plant extracts agroinfiltrated with PVX.
In 2017, Chahardoli et al. demonstrated the expression of a chimeric hLf peptide in tobacco Hairy Roots (HR) and investigated the antibacterial activity of this peptide. The nucleotide sequence encoding the Lfchimera gene consisted of the sequences of lactoferricin and lactoferrampin peptides and endoplasmic reticulum (ER) retention signal peptide; the Lfchimera gene was placed under the control of the CaMV35S promoter. Later, Chahardoli et al. (2018) reported A. tumefaciens-mediated transformation of tobacco plants with the same Lfchimera gene as in Chahardoli et al. (2017). Antibacterial activity of total protein extracts and purified Lfchimera was demonstrated against two species of clinical bacteria (E. coli and S. aureus) and two phytopathogenic bacteria (E. amylovora and R. solanacearum). Therefore, this work showed that the expression of a functionally active chimeric peptide derived from Lf in tobacco plants did not affect the growth and morphology of transgenic plants (Chahardoli et al. 2018).
The works by (Takase et al. 2005; Li et al. 2004; Chahardoli et al. 2018) showed that the transformation of O. sativa, N. tabacum and N. benthamiana plants with fragments of the human lactoferrin gene did not alter the morphology of transgenic plants, in contrast to studies in which rice and alfalfa plants were transformed with the full-length hLf gene (Stefanova et al. 2013a, b; Takase et al. 2005). At the same time, Takase et al. (2005) showed that the expression of neither hLf nor hLfN gene changed the resistance of rice plants to the fungal pathogen (P. oryzae). Moreover, Li et al. (2004) showed that the expression of hLfN did not alter the post-inoculation resistance of N. benthamiana to PVX. Nevertheless, Takase et al. (2005) and Chahardoli et al. (2018) showed that the expression of the Lfchimera and hLfN could enhance the resistance of transgenic plants to bacterial pathogens (P. plantarii, E. amylovora and R. solanacearum) (Table 1). Thus, we can conclude that transformation with the full-length human lactoferrin gene (Zhang et al. 1998; Stefanova et al. 2013a, b; Lee et al. 2002; Buziashvili et al. 2020a, b; Takase et al. 2005; Malnoy et al. 2003; Nguyen et al. 2011; Han et al. 2012) more effectively increases plant resistance to phytopathogens than transformation with the fragments of the lactoferrin gene (Takase et al. 2005; Li et al. 2004).
It should be noted that the influence of lactoferrin expression on plant physiology was thoroughly studied by Kumar et al. (2013). The effect of hLf expression on iron homeostasis was assessed by measuring the content of iron, chlorophyll, phenols, flavonoids, proanthocyanidins and anthocyanins, and by assessing the levels of expression of the genes involved in iron homeostasis. Overall, the results of this study indicate iron deficiency in transgenic tobacco plants expressing hLf. Such effects are minor compared to enhanced plant immunity as a positive outcome of Lf expression, which can generally increase the productivity of transgenic plants.
Transformation of plants with the full-length bovine lactoferrin gene
To investigate the ability of bovine lactoferrin to increase the resistance of transgenic plants to the fungal phytopathogen Rhizoctonia solani, the causative agent of root rot and seedling damping-off diseases, Nguyen et al. (2011) performed Agrobacterium-mediated transformation of N. tabacum and A. thaliana using the bLf gene driven by the CaMV35S promoter. In general, the results of all biotests indicate that the expression of bLf in the transgenic A. thaliana and N. tabacum plants enhances their resistance to R. solani. The authors suppose that such effect might be the result of Lf-induced senescence of the leaves from transgenic plants. Thus, as the authors (Nguyen et al. 2011) concluded, the heterologous expression of bovine lactoferrin could be applied to protect economically valuable plants from diseases caused by R. solani.
In order to enhance the resistance of wheat cultivar Bobwhite susceptible to fusarium head blight, Han et al. (2012) performed a transformation with the bovine lactoferrin gene under the control of Adenine Methyltransferase Promoter (AMTP). BLf content differed in leaves (0.52% of TSP) and glumes (0.11% of TSP). The resistance of transgenic plants to Fusarium graminearum was confirmed in both in vitro and in vivo assays. The infection rates were estimated at 14–46% for transgenic lines, 82% for untransformed Bobwhite line (negative control) and 27% for resistant cultivar ND 2710 (positive control). The results show an increased resistance of plants expressing bLf to FHB. The authors note that the level of resistance was stable within the line and correlated with the levels of bLf expression. The expression of lactoferrin did not alter the morphology of transgenic plants. The results suggest that the bLf gene is a promising means for increasing the resistance of cereals to phytopathogenic fungi (Han et al. 2012).
Transformation of pear (Pyrus communis L.) with the bovine lactoferrin gene (bLf) to increase the resistance to fire blight disease was carried out in 2003 by Malnoy et al. It was carried out with the bLf gene under the control of CaMV35S promoter. All transgenic lines had growth retardation which was less significant in greenhouse conditions. The authors suggest that this effect could be either due to the influence of lactoferrin expression and its iron-chelating properties or to the somaclonal variability of transgenic lines. The enhancement of the resistance of transgenic plants to Erwinia amylovora, a siderophore-carrying bacteria causing fire blight disease, and to several other economically important pear pathogens, namely, Pseudomonas syringae pv. syringae causing bacterial pear blast and A.tumefaciens, the causative agent of crown gall disease, was confirmed with the use of in vitro and in vivo biotests. The results of in vitro assay also showed that the bacteriostatic effect of lactoferrin on E. amylovora is exerted mainly through its ferrum-chelating activity. The results of this study show that despite some minor effects such as growth retardation, the expression of bLf causes considerable pathogen-protecting effects on transgenic pear (Malnoy et al. 2003). Later, in 2011, similar research was carried out by Djennane et al. (2011) Transformation of pear cv. Passe Crassane with the gene of another ferrum-binding protein, the ferritin gene of pea, was performed to enhance pear resistance to E. amylovora. Biotests showed that the transgenic pear plants expressing the ferritin gene had no resistance to E. amylovora. Thus, among the two ferrum-chelating proteins, lactoferrin reveals a stronger antibacterial activity than ferritin.
Thus, growth retardation due to transformation with the bLf gene was detected only in transgenic pear plants (Malnoy et al. 2003), while the morphology of transgenic tobacco, Arabidopsis and wheat plants was similar to control (Nguyen et al. 2011; Han et al. 2012). According to Han et al. (2012), the bLf gene was expressed under the control of the AMTP promoter, and the lactoferrin content was at a similar level (0.11–0.52% of TSP) as in the work in which lactoferrin genes were driven by the CaMV35S promoter (Table 1). It can be concluded therefore, that no significant differences in the levels of lactoferrin expression were observed in transgenic plants expressing the full-length bovine lactoferrin gene under the control of different types of promoters (Table 1). However, the correlation between the content of the lactoferrin proteins or lactoferrin-derived peptides produced in transgenic plants and the levels of their resistance to phytopathogens needs further studies.
Transformation of plants with the bovine lactoferrin-derived gene
There is evidence of transformation of N. tabacum by the gene of lactoferricin B (bLfcin), an antimicrobial N-terminal peptide obtained by acidic pepsin hydrolysis of bovine lactoferrin (Fukuta et al. 2012). This gene was fused to the nucleotide sequence of a signal peptide from a tobacco pathogenesis-related protein (PR-1) to target its secretion into the intercellular apoplast region and enhance the pathogen-induced immune response and is controlled by CaMV35S promoter. It was established that the obtained transgenic plants were more resistant to the bacterial pathogen Pseudomonas syringae pv. tabaci (the causative agent of wildfire disease of tobacco) and the fungal pathogen Botrytis cinerea (the causal agent of grey mold in many plant species) than the control plants. Thus, the results of Fukuta et al. (2012) show that transformation of N. tabacum with the bLfcin gene enhances resistance of transgenic plants to bacterial (P. syringae) and fungal (B. cinerea) pathogens and does not alter the morphology of the transgenic plants (Table 1).
Conclusions
In general, plant protection strategies against phytopathogens are based on the expression of certain genes, which leads to various responses, such as HR (in the case of SAR), cell wall modification, synthesis of antimicrobial compounds, e.g. AMP, or iron sequestration (in the case of ISR). The transfer of genes encoding antimicrobial peptides or proteins into plant genomes is a promising means of protecting plants against various diseases. One such protein is lactoferrin, a well-studied mammalian iron-binding protein with strong antimicrobial and immunomodulatory activity, and low allergenicity. Due to the various beneficial properties of lactoferrin, its expression in the plant genome could reinforce the resistance of transgenic plants to a wide range of different bacterial (X. campestris, P. syringae, R. solanacearum, C. michiganensis, E. amylovora, P. plantarii, A. tumefaciens), fungal (R. solani, B. cinerea, P. infestans, F. graminearum), and some viral pathogens such as Rice Dwarf Virus. This approach has been successfully applied to different species of agricultural and model plants, namely A. thaliana, N. benthamiana, N. tabacum, S. lycopersicum, M. sativa, O. sativa, S. tuberosum and T. aestivum. Given all the benefits of transforming plants with lactoferrin genes, we can conclude that this biotechnological approach is a very promising way to achieve effective protection of crops from various diseases.
References
Abdallah FB, El Hage Chahine JM (2000) Transferrins: iron release from lactoferrin. J Mol Biol 303:255–266. https://doi.org/10.1006/jmbi.2000.4101
Abdelbacki AM, Taha SH, Sitohy MZ, Dawood AIA, Hamid MMA, Rezk AA (2010) Inhibition of Tomato yellow leaf curl virus (TYLCV) using whey proteins. Virol J. https://doi.org/10.1186/1743-422X-7-26
Abdelghany MMA, Kurikawa M, Watanabe M, Matsui H, Yamamoto M, Ichinose Y, Toyoda K, Kouzai Y, Noutoshi Y (2022) Surveillance of pathogenicity of Rhizoctonia solani Japanese isolates with varied anastomosis groups and subgroups on Arabidopsis thaliana. Life. https://doi.org/10.3390/life12010076
Adleranova L, Bartoskova A, Faldyna M (2008) Lactoferrin: a review. Vet Med 53:457–468. https://doi.org/10.17221/1978-VETMED
Ali I, Khan MS, Mustafa G, Sultan N, Iqbal Z, Shafiq M (2018) Chapter 4. Resistance strategies against microbial plant pathogens. In: Current research in microbiology. Open access ebooks, pp 1–23
Andersen EJ, Ali S, Byamukama E, Yen Y, Nepal MP (2018) Disease resistance mechanisms in plants. Genes. https://doi.org/10.3390/genes9070339
Bennett RM, Bagby GC, Davis J (1981) Calcium-dependent polymerization of lactoferrin. Biochem Biophys Res Commun 101:88–95. https://doi.org/10.1016/S0006-291X(81)80014-9
Berlutti F, Pantanella F, Natalizi T, Frioni A, Paesano R, Polimeni A, Valenti P (2011) Antiviral properties of lactoferrin—a natural immunity molecule. Molecules 16:6992–7018. https://doi.org/10.3390/molecules16086992
Bielecka M, Cichosz G, Czeczot H (2022) Antioxidant, antimicrobial and anticarcinogenic activities of bovine milk proteins and their hydrolysates—a review. Int Dairy J. https://doi.org/10.1016/j.idairyj.2021.105208
Boddy L (2016) Pathogens of autotrophs. In: Watkinson SC, Money N, Boddy L (eds) The fungi, 3rd edn. Academic Press, Cambridge, p 466
Bruce TJ (2012) GM as a route for delivery of sustainable crop protection. J Exp Bot 63:537–541. https://doi.org/10.1093/jxb/err281
Bruni N, Capucchio MT, Biasibetti E, Pessione E, Cirrincione S, Giraudo L, Corona A, Dosio F (2016) Antimicrobial activity of lactoferrin-related peptides and applications in human and veterinary medicine. Molecules. https://doi.org/10.3390/molecules21060752
Buziashvili A, Cherednichenko L, Kropyvk S, Blume Y, Yemets A (2020a) Obtaining transgenic potato plants expressing the human lactoferrin gene and analysis of their resistance to phytopathogens. Cytol Genet 54:179–188. https://doi.org/10.3103/S0095452720030020
Buziashvili A, Cherednichenko L, Kropyvko S, Yemets A (2020b) Transgenic tomato lines expressing human lactoferrin show increased resistance to bacterial and fungal pathogens. Biocatal Agricult Biotechnol. https://doi.org/10.1016/j.bcab.2020.101602
Ceasar SA, Ignacimuthu S (2012) Genetic engineering of crop plants for fungal resistance: role of antifungal genes. Biotechnol Lett 34:995–1002. https://doi.org/10.1007/s10529-012-0871-1
Cerda R, Avelino J, Gary C, Tixier P, Lechevallier E, Allinne C (2017) Primary and secondary yield losses caused by pests and diseases: Assessment and modeling in coffee. PLoS ONE. https://doi.org/10.1371/journal.pone.0169133
Chahardoli M, Fazeli A, Ghabooli M (2017) Recombinant production of bovine Lactoferrin-derived antimicrobial peptide in tobacco hairy roots expression system. Plant Physiol Biochem 123:414–421. https://doi.org/10.1016/j.plaphy.2017.12.037
Chahardoli M, Fazeli A, Niazi A, Ghabooli M (2018) Recombinant expression of LFchimera antimicrobial peptide in a plant-based expression system and its antimicrobial activity against clinical and phytopathogenic bacteria. Biotechnol Biotechnol Equip 32:714–723. https://doi.org/10.1080/13102818.2018.1451780
Chaloner TM, Gurr SJ, Bebber DP (2021) Plant pathogen infection risk tracks global crop yields under climate change. Nat Clim Chang 11:710–715. https://doi.org/10.1038/s41558-021-01104-8
Choudhary DK, Prakash A, Johri BN (2007) Induced systemic resistance (ISR) in plants: mechanism of action. Indian J Microbiol 47:289–297. https://doi.org/10.1007/s12088-007-0054-2
Conrath U (2006) Systemic acquired resistance. Plant Signal Behav 1:179–184. https://doi.org/10.4161/psb.1.4.3221
de Vleesschauwer D, Höfte M (2009) Rhizobacteria-induced systemic resistance. Adv Bot Res 51:223–281. https://doi.org/10.1016/S0065-2296(09)51006-3
Djennane S, Cesbron C, Sourice S, Cournol R, Dupuis F, Eychenne M, Loridon K, Chevreau E (2011) Iron homeostasis and fire blight susceptibility in transgenic pear plants overexpressing a pea ferritin gene. Plant Sci 180:694–701. https://doi.org/10.1016/j.plantsci.2011.01.015
Durrant WE, Dong X (2004) Systemic acquired resistance. Annu Rev Phytopathol 42:185–209. https://doi.org/10.1146/annurev.phyto.42.040803.140421
Emeriewen OF, Wöhner T, Flachowsky H, Peil A (2019) Malus Hosts-Erwinia amylovora interactions: strain pathogenicity and resistance mechanisms. Front Plant Sci. https://doi.org/10.3389/fpls.2019.00551
Fernandes KE, Carter DA (2017) The antifungal activity of lactoferrin and its derived peptides: mechanisms of action and synergy with drugs against fungal pathogens. Front Microbiol. https://doi.org/10.3389/fmicb.2017.00002
Fukuta S, Kawamoto K, Mizukami Y, Yoshimura Y, Ueda J, Kanbe M (2012) Transgenic tobacco plants expressing antimicrobial peptide bovine lactoferricin show enhanced resistance to phytopathogens. Plant Biotech 29:383–389. https://doi.org/10.5511/plantbiotechnology.12.0619a
Garfinkel AR, Coats KP, Sherry DL, Chastagner GA (2019) Genetic analysis reveals unprecedented diversity of a globally-important plant pathogenic genus. Sci Rep. https://doi.org/10.1038/s41598-019-43165-y
Giansanti F, Panella G, Leboffe L, Antonimi G (2016) Lactoferrin from milk: nutraceutical and pharmacological properties. Pharmaceuticals. https://doi.org/10.3390/ph9040061
Glazebrook J (2005) Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Annu Rev Phytopathol 43:205–227. https://doi.org/10.1146/annurev.phyto.43.040204.135923
Grant MR, Kazan K, Manners JM (2013) Exploiting pathogens’ tricks of the trade for engineering of plant disease resistance: challenges and opportunities. Microb Biotechnol 6:212–222. https://doi.org/10.1111/1751-7915.12017
Gruden Š, Poklar Ulrih N (2021) Diverse mechanisms of antimicrobial activities of lactoferrins, lactoferricins, and other lactoferrin-derived peptides. Int J Mol Sci. https://doi.org/10.3390/ijms222011264
Hakkansson A, Zhivotovsky B, Orrenius S, Sabharwal H, Svanborg C (1995) Apoptosis induced by a human milk protein. Proc Nat Acad Sci Unit St Am 92:8064–8068. https://doi.org/10.1073/pnas.92.17.8064
Han J, Lakshman DK, Galvez LC, Mitra S, Baenziger PS, Mitra A (2012) Transgenic expression of lactoferrin imparts enhanced resistance to head blight of wheat caused by Fusarium graminearum. Plant Biol 12:33–42. https://doi.org/10.1186/1471-2229-12-33
Han GZ (2019) Origin and evolution of the plant immune system. New Phytol 222(1):70–83. https://doi.org/10.1111/nph.15596
Henry E, Yadeta KA, Coaker G (2013) Recognition of bacterial plant pathogens: local, systemic and transgenerational immunity. New Phytol 199:908–915. https://doi.org/10.1111/nph.12214
Jacobs JM, Milling A, Mitra RM, Hogan CS, Ailloud F, Prior P, Allen C (2013) Ralstonia solanacearum requires PopS, an ancient AvrE-family effector, for virulence and to overcome salicylic acid-mediated defenses during tomato pathogenesis. Mbio. https://doi.org/10.1128/mBio.00875-13
Jahani S, Shakiba A, Jahani L (2015) The antimicrobial effect of lactoferrin on gram-negative and gram-positive bacteria. Int J Infect. https://doi.org/10.17795/iji27594
Jones JDG, Dangl JL (2006) The plant immune system. Nature 444:323–329. https://doi.org/10.1038/nature05286
Jung Y, Kang K (2014) Application of antimicrobial peptides for disease control in plants. Plant Breed Biotech 2:1–13. https://doi.org/10.9787/PBB.2014.2.1.001
Kell DB, Heyden EL, Pretorius E (2020) The biology of lactoferrin, an iron-binding protein that can help defend against viruses and bacteria. Front Immunol. https://doi.org/10.3389/fimmu.2020.01221
Kraepiel Y, Barny MA (2016) Gram-negative phytopathogenic bacteria, all hemibiotrophs after all? Mol Plant Pathol 17:313–316. https://doi.org/10.1111/mpp.12345
Kumar V, Gill T, Grover S, Ahuja PS, Yadav SK (2013) Influence of human lactoferrin expression on iron homeostasis, flavonoids, and antioxidants in transgenic tobacco. Mol Biotechnol 53:118–128. https://doi.org/10.1007/s12033-012-9495-x
Lahoz E, Pisacane A, Iannaccone M, Palumbo D, Capparelli R (2008) Fungistatic activity of iron-free bovin lactoferrin against several fungal plant pathogens and antagonists. Nat Prod Res 22:955–961. https://doi.org/10.1080/14786410701650253
Lakshman DK, Natarajan S, Mandal S, Mitra A (2013) Lactoferrin derived resistance against plant pathogens in transgenic plants. J Agric Food Chem 61:11730–11735. https://doi.org/10.1021/jf400756t
Lee TJ, Coyne PP, Clemente TE, Mitra A (2002) Partial resistance to bacterial wilt in transgenic tomato plants expressing antibacterial lactoferrin gene. J Am Soc Hort Sci 127:158–168. https://doi.org/10.21273/JASHS.127.2.158
Li Y, Geng Y, Song H, Zheng G, Huan L, Qiu B (2004) Expression of a human lactoferrin N-lobe in Nicotiana benthmiana with potato virus X-based agroinfection. Biotechnol Lett 26:953–947. https://doi.org/10.1023/B:BILE.0000030038.27358.20
Li J, Hu S, Jian W, Yang X (2021) Plant antimicrobial peptides: structures, functions, and applications. Bot Stud. https://doi.org/10.1186/s40529-021-00312-x
van Loon LC, Bakker PAHM (2005) Induced systemic resistance as a mechanism of disease suppression by rhizobacteria. In: Siddiqui ZA (ed) PGPR: Biocontrol and Biofertilization. Springer, Dordrecht, pp 39–66. https://doi.org/10.1007/1-4020-4152-7_2.
López-García B, San Segundo B, Coca M (2012) Chapter 13. Antimicrobial peptides as a promising alternative for plant disease protection. In: Small wonders: peptides for disease control. ACS Symposium series. pp 263–294. https://doi.org/10.1021/bk-2012-1095.ch013
Malnoy M, Venisse JS, Brisset MN, Chevreau E (2003) Expression of bovine lactoferrin cDNA confers resistance to Erwinia amylovora in transgenic pear. Molec Breed 12:231–244. https://doi.org/10.1023/A:1026365311067
Mantel C, Miyazawa K, Broxmeyer HE (1994) Physical characteristics and polymerization during iron saturation of lactoferrin, a myelopoietic regulatory molecule with suppressor activity. Adv Exp Med Biol 357:121–132. https://doi.org/10.1007/978-1-4615-2548-6_12
Marcos JF, Muñoz A, Pérez-Payá E, Misra S, López-García B (2008) Identification and rational design of novel antimicrobial peptides for plant protection. Annu Rev Phytopathol 46:273–301. https://doi.org/10.1146/annurev.phyto.121307.094843
Mas Muniroh MN (2018) Different approaches of combating bacterial canker in tomato: in pursuit of resistance. Dissertation, Wageningen University
McDonald BA, Stukenbrock EH (2016) Rapid emergence of pathogens in agro-ecosystems: global threats to agricultural sustainability and food security. Philos Trans R Soc Lond B Biol Sci 371:20160026. https://doi.org/10.1098/rstb.2016.0026
Mitra A, Zhang Z (1994) Expression of a human lactoferrin cDNA in tobacco cells produces antibacterial protein(s). Plant Physiol 106:977–981
Moosa A, Farzand A, Sahi ST, Khan SA (2017) Transgenic expression of antifungal pathogenesis-related proteins against phytopathogenic fungi – 15 years of success. Isr J Plant Sci 65:38–54. https://doi.org/10.1080/07929978.2017.1288407
Muñoz A, Marcos JF (2006) Activity and mode of action against fungal phytopathogens of bovine lactoferricin-derived peptides. J Appl Microbiol 101:1199–1207. https://doi.org/10.1111/j.1365-2672.2006.03089.x
Nguyen TC, Lakshman DK, Han J, Galvez LC, Mitra A (2011) Transgenic plants expressing antimicrobial lactoferrin protein are resistant to a fungal pathogen. J Plant Mol Biol Biotechnol 2:1–8
Nowicki M, Foolad MR, Nowakowska M, Kozik EU (2011) Potato and tomato late blight caused by Phytophthora infestans: an overview of pathology and resistance breeding. Plant Dis 96:4–17. https://doi.org/10.1094/PDIS-05-11-0458
Oda H, Wakabayashi H, Tanaka M, Yamauchi K, Sugita C, Yoshida H, Abe F, Sonoda T, Kurokawa M (2021) Effects of lactoferrin on infectious diseases in Japanese summer: a randomized, double-blinded, placebo-controlled trial. J Microbiol Immunol Infect 54:566–574
Osusky M, Zhou G, Osuska L, Hancock RE, Kay WW, Misra S (2000) Transgenic plants expressing cationic peptide chimeras exhibit broad-spectrum resistance to phytopathogens. Nat Biotechnol 18:1162–1166. https://doi.org/10.1038/81145
Owen W, Punja ZK (2010) Genetic engineering for increasing fungal and bacterial disease resistance in crop plants. GM Crops 1:199–206. https://doi.org/10.4161/gmcr.1.4.13225
Patil VU, Gopa J, Singh BP (2012) Improvement for bacterial wilt resistance in potato by conventional and biotechnological approaches. Agric Res 1:299–316. https://doi.org/10.1007/s40003-012-0034-6
Peng Y, Li SJ, Yan J, Tang Y, Cheng JP, Gao AJ, Yao X, Ruan JJ, Xu BL (2021) Research progress on phytopathogenic fungi and their role as biocontrol agents. Front Microbiol. https://doi.org/10.3389/fmicb.2021.670135
Pieterse CM, Zamioudis C, Berendsen RL, Weller DM, van Wees SC, Bakker PA (2014) Induced systemic resistance by beneficial microbes. Annu Rev Phytopathol 52:347–375. https://doi.org/10.1146/annurev-phyto-082712-102340
Rao HCY, Jayabhaskaran C, Kamalraj S, Parthasarathy R, Mondal S, Sundararaj R, Kumar S (2021) Chapter 4—exploring the molecular signatures of host–pathogen interactions in plant diseases: conflict and cooperation. In: Ajay K, Samir D (eds) Food security and plant disease management. Woodhead Publishing, Sawston, pp 63–74. https://doi.org/10.1016/B978-0-12-821843-3.00003-9
Rauwane ME, Ogugua UV, Kalu CM, Ledwaba LK, Woldesemayat AA, Ntushelo K (2020) Pathogenicity and virulence factors of Fusarium graminearum including factors discovered using next generation sequencing technologies and proteomics. Microorganisms. https://doi.org/10.3390/microorganisms8020305
Raynaud X, Nunan N (2014) Spatial ecology of bacteria at the microscale in soil. PLoS ONE. https://doi.org/10.1371/journal.pone.0087217
Razzaq A, Kaur P, Akhter N, Wani HS, Fozia S (2021) Next-generation breeding strategies for climate-ready crops. Front Plant Sci. https://doi.org/10.3389/fpls.2021.620420
Romera FJ, García MJ, Lucena C, Martínez-Medina A, Aparicio MA, Ramos J, Alcántara E, Angulo M, Pérez-Vicente R (2019) Induced systemic resistance (ISR) and Fe deficiency responses in dicot plants. Front Plant Sci. https://doi.org/10.3389/fpls.2019.00287
Rommens CM, Kishore GM (2000) Exploiting the full potential of disease-resistance genes for agricultural use. Curr Opinion Biotechnol 11:120–125. https://doi.org/10.1016/s0958-1669(00)00083-5
Salmon V, Legrand D, Slomianny MC, el Yazidi I, Spik G, Gruber V, Bournat P, Olagnier B, Mison D, Theisen M, Mérot B (1998) Production of human lactoferrin in transgenic tobacco plants. Protein Expr Purif 13:127–135. https://doi.org/10.1006/prep.1998.0886
Savary S, Ficke A, Aubertot J-N, Hollier C (2012) Crop losses due to diseases and their implications for global food production losses and food security. Food Secur 4:519–537. https://doi.org/10.1007/s12571-012-0200-5
Shuping DSS, Eloff JN (2017) The use of plants to protect plants and food against fungal pathogens: a review. Afr J Tradit Complement Altern Med 14:120–127. https://doi.org/10.21010/ajtcam.v14i4.14
Sinha R, Shukla P (2019) Antimicrobial peptides: recent insights on biotechnological interventions and future perspectives. Protein Pept Lett 26:79–87. https://doi.org/10.2174/0929866525666181026160852
Sinha M, Kaushik S, Kaur P, Sharma S, Singh TP (2013) Antimicrobial lactoferrin peptides: the hidden players in the protective function of a multifunctional protein. Int J Pept 12:390–230. https://doi.org/10.1155/2013/390230
Spik G, Strecker G, Fournet B, Bouquelet S, Montreuil J (1982) Primary structure of the glycans from human lactotransferrin. Eur J Biochem 121:413–419
Stefanova G, Vlahova M, Atanassov A (2008) Production of recombinant human lactoferrin from transgenic plants. Biol Plant 52:423–428. https://doi.org/10.1007/s10535-008-0086-4
Stefanova G, Slavov S, Gecheff K, Vlahova M, Atanassov A (2013a) Expression of recombinant human lactoferrin in transgenic alfalfa plants. Biol Plant 57:457–464. https://doi.org/10.1007/s10535-013-0305-5
Stefanova G, Vassileva V, Vlahova M (2013b) Human lactoferrin changes leaf morphology and pathogen resistance of Medicago sativa L. Bulg J Agric Sci 19:706–713
Takase K, Hagiwara K, Onodera H, Nishizawa Y, Ugaki M, Omura T, Numata S, Akutsu K, Kumura H, Shimazaki K (2005) Constitutive expression of human lactoferrin and its N-lobe in rice plants to confer disease resistance. Biochem Cell Biol 83:239–249. https://doi.org/10.1139/o05-022
Tanasienko IV, Yemets AI, Pirko YV, Korhkovyy VI, Abumhadi N, Blume YB (2011) Generation of transgenic barley lines producing human lactoferrin using mutant alpha-tubulin gene as the selective marker. Cyt Genet 45:3–10. https://doi.org/10.3103/S0095452711010026
Teng PS, Shane WW, MacKenzie DR (1984) Crop losses due to plant pathogens. Crit Rev Plant Sci 2:21–47. https://doi.org/10.1080/07352688409382187
Vallad GE, Goodman RM (2004) Systemic acquired resistance and induced systemic resistance in conventional agriculture. Crop Sci 44:1920–1934. https://doi.org/10.2135/cropsci2004.1920
Varshney RK, Bohra A, Yu J, Graner A, Zhang Q, Sorrells ME (2021) Designing future crops: genomics-assisted breeding comes of age. Trends Plant Sci 26:631–649. https://doi.org/10.1016/j.tplants.2021.03.010
Vlot AC, Sales JH, Lenk M, Bauer K, Brambilla A, Sommer A, Chen Y, Wenig M, Nayem S (2020) Systemic propagation of immunity in plants. New Phytol. https://doi.org/10.1111/nph.16953
Wakabayashi H, Oda H, Yamauchi K, Abe F (2014) Lactoferrin for prevention of common viral infections. Infect Chemother 20:666–671. https://doi.org/10.1016/j.jiac.2014.08.003
Wang R, Han Z, Ji R, Xiao Y, Si R, Guo F, He J, Hai L, Ming L, Yi L (2020a) Antibacterial activity of trypsin-hydrolyzed Camel and Cow whey and their fractions. Animals. https://doi.org/10.3390/ani10020337
Wang Y, Wang P, Wang H, Luo Y, Wan L, Jiang M, Chu Y (2020b) Lactoferrin for the treatment of COVID-19 (review). Exp Ther Med. https://doi.org/10.3892/etm.2020.9402
Yemets AI, Tanasienko IV, Krasylenko YA, Blume YB (2014) Plant-based biopharming of recombinant human lactoferrin. Cell Biol Int 38:989–1002. https://doi.org/10.1002/cbin.10304
Zhang Z, Coyne DP, Vidaver AK, Mitra A (1998) Expression of human lactoferrin cDNA confers resistance to Ralstonia solanacearum in transgenic tobacco plants. Phytopathology 88:730–734. https://doi.org/10.1094/PHYTO
Zhang J, Coaker G, Zhou J-M, Dong X (2020) Plant immune mechanisms: from reductionistic to holistic points of view. Mol Plant 13:1358–1378. https://doi.org/10.1016/j.molp.2020.09.007
Zhang Y, Lu C, Zhang J (2021) Lactoferrin and its detection methods: a review. Nutrients. https://doi.org/10.3390/nu13082492
Acknowledgements
The authors are grateful to Dr. V. Kyrylenko (Institute of Food Biotechnology and Genomics, National Academy of Sciences of Ukraine, Kyiv) for final improving the English version of the manuscript.
Funding
This work was financially supported by the National Academy of Sciences of Ukraine under the budget theme”Study of the response of plants to abiotic and biotic factors at the cellular and genetic levels to improve plant adaptive properties against the negative effects of climate change” (state registration number #0117U000909, 2017–2021).
Author information
Authors and Affiliations
Contributions
AB—Analyzed and interpreted the data; Wrote the paper. AY—Reviewed and analyzed the manuscript contents; Made critical remarks and the manuscript corrections. All authors have approved the final manuscript version.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Buziashvili, A., Yemets, A. Lactoferrin and its role in biotechnological strategies for plant defense against pathogens. Transgenic Res 32, 1–16 (2023). https://doi.org/10.1007/s11248-022-00331-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11248-022-00331-9