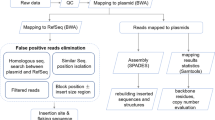
Abstract
Genomic insertions and flanking regions of transgenes in host genomes constitute a critical component of precise molecular characterization and event-specific detection, which are required in the development and assessment for regulatory approval of genetically modified (GM) crops. Previously, we reported three transgenic soybean events harboring the inverted repeats of the soybean mosaic virus NIb (nuclear inclusion b) gene, exhibiting significantly enhanced resistance to multiple Potyvirus strains. To facilitate safety assessment and event-specific detection, we identified the transgene insertion sites and flanking sequences of the events L120, L122, and L123 using whole-genome sequencing. More than 14.48 Gb sequence data (13 × coverage) were generated using the Illumina HiSeq Xten platform for each event. The sequence reads corresponding to boundaries of inserted T-DNA, and associated native flanking sequences were identified by bioinformatic comparison with the soybean reference genome (Wm82.a2.v1) and the transformation vector sequence. The results indicated that two T-DNA insertions occurred in L120, on Chr07 and Chr13, while L122 and L123 showed single insertions, on Chr02 and Chr06, respectively. Based on the flanking sequences of the inserted T-DNA, the event-specific detection for each event was established using specific PCR primers, and PCR amplification followed by sequencing of PCR products further confirmed the putative insertion loci and flanking regions in the transgenic lines. Our results demonstrate the efficacy and robustness of whole-genome sequencing in identifying the genomic insertions and flanking regions in GM crops. Moreover, the characterization of insertion loci and the establishment of event-specific detection will facilitate the application and development of broad-spectrum virus-resistant transgenic soybean cultivars.



Similar content being viewed by others
References
Ajay SS, Parker SC, Abaan HO, Fajardo KV, Marqulies EH (2011) Accurate and comprehensive sequencing of personal genomes. Genome Res 21(9):1498–1505
Babekova R, Funk T, Pecoraro S, Engel KH, Busch U (2009) Development of an event-specific real-time PCR detection method for the transgenic Bt rice line KMD1. Eur Food Res Technol 228(5):707–716
Campbell PJ, Stephens PJ, Pleasance ED, O’Meara S, Li H, Santarius T, Stebbings LA, Leroy C, Edkins S, Hardy C, Teague JW, Menzies A, Goodhead I, Turner DJ, Clee CM, Quail MA, Cox A, Browm C, Durbin R, Hurles ME, Edwards PA, Bignell G, Stratton MR, Futreal PA (2008) Identification of somatically acquired rearrangements in cancer using genome-wide massively parallel paired-end sequencing. Nat Genet 40(6):722–729
Chen SY, Jin WZ, Wang MY, Zhang F, Zhou J, Jia QJ, Wu YR, Liu FY, Wu P (2003) Distribution and characterization of over 1000 T-DNA tags in rice genome. Plant J 36(1):105–113
Codex Alimentarius (2003) Guideline for the conduct of food safety assessment of foods derived from recombinant-DNA plants. Codex Alimentarius Commission, Joint FAO/WHO Food Standards Programme, Food and Agriculture Organization of the United Nations, Rome, Italy
Dubose AJ, Lichtenstein ST, Narisu N, Bonnycastle LL, Swift AJ, Chines PS, Collins FS (2013) Use of microarray hybrid capture and next-generation sequencing to identify the anatomy of a transgene. Nucleic Acids Res 41(6):e70
European Food Safety Authority [EFSA] (2010) Guidance on the environmental risk assessment of genetically modifified plants. EFSA J 8:1879
Guo B, Yong G, Hong H, Qiu LJ (2016) Identification of genomic insertion and flanking sequence of G2-EPSPS and GAT transgenes in soybean using whole genome sequencing method. Front Plant Sci 7:1009
Guo L, Liang SJ, Zhang ZY, Liu H, Wang SW, Pan KH, Xu J, Ren X, Pei SR, Yang GP (2019) Genome assembly of Nannochloropsis oceanica provides evidence of host nucleus overthrow by the symbiont nucleus during speciation. Commun Biol 2:249
Hormozdiari F, Hajirasouliha I, Mcpherson A, Eichler EE, Sahinalp C (2011) Simultaneous structural variation discovery among multiple paired-end sequenced genomes. Genome Res 21(12):2203–2212
Inagaki S, Henry IM, Lieberman MC, Comai L (2015) High-throughput analysis of T-DNA location and structure using sequence capture. PLoS ONE 10(10):e0139672
Kok E, Pedersen J, Onori R, Sowa S, Schauzu M, Schrijver AD, Teeri TH (2014) Plants with stacked genetically modified events: to assess or not to assess? Trends Biotechnol 32(2):70–73
Kovalic D (2012) The use of next generation sequencing and junction sequence analysis bioinformatics to achieve molecular characterization of crops improved through modern biotechnology. Plant Genome 5(3):149–163
Langmead B, Salzberg SL (2012) Fast gapped-read alignment with Bowtie 2. Nat Methods 9:357–359
Latham JR, Wilson AK (2006) Steinbrecher RA (2006) The mutational consequences of plant transformation. J Biomed Biotechnol 2:25376
Lepage E, Zampini E, Boyle B, Brisson N (2013) Time and cost-efficient identification of T-DNA insertion sites through targeted genomic sequencing. PLoS ONE 8(8):e70912
Liang CJ, van Dijk JP, Scholtens IM, Staats M, Prins TW, Voorhuijzen MM, da Silva AM, Arisi AC, den Dunnen JI, Kok EJ (2014) Detecting authorized and unauthorized genetically modified organisms containing vip3A by real-time PCR and next-generation sequencing. Anal Bioanal Chem 406(11):2603–2611
Liu YG, Mitsukawa N, Oosumi T, Whittier RF (1995) Efficient isolation and mapping of Arabidopsis thaliana T-DNA insert junctions by thermal asymmetric interlaced PCR. Plant J 8(3):457–463
Milcamps A, Rabe S, Cade R, De Framond AJ, Henriksson P, Kramer V, Lisboa D, Pastor-Benito S, Willits MG, Lawrence D, van den Eede G (2009) Validity assessment of the detection method of maize event Bt10 through investigation of its molecular structure. J Agric Food Chem 57(8):3156–3163
Nacry P, Camilleri C, Courtial B, Caboche M, Bouchez D (1998) Major chromosomal rearrangements induced by T-DNA transformation in Arabidopsis. Genetics 149(2):641–650
Ochman H, Gerber AS, Hartl DL (1998) Genetic applications of an inverse polymerase chain reaction. Genetics 120(3):621–623
O’Malley RC, Ecker JR (2010) Linking genotype to phenotype using the Arabidopsis unimutant collection. Plant J 61(6):928–940
Park D, Kim D, Jang G, Lim JS, Shin YJ, Kim J, Seo MS, Park SH, Kim JK, Kwon TH, Choi IY (2015) Efficiency to discovery transgenic loci in GM rice using next generation sequencing whole genome re-sequencing. Genomics Inform 13(3):81
Porebski S, Bailey LG, Baum BR (1997) Modification of a CTAB DNA extraction protocol for plants containing high polysaccharide and polyphenol components. Plant Mol Biol Rep 15(1):8–15
Siddique K, Wei JJ, Li R, Zhang DB, Shi JX (2019) Identification of T-DNA insertion site and flanking sequence of a genetically modified maize event IE09S034 using next-generation sequencing technology. Mol Biotechnol 61(9):694–702
Spalinskas R, Van den Bulcke M, Van den Eede G, Milcamps A (2013) LT-RADE: an efficient user-friendly genome walking method applied to the molecular characterization of the insertion site of genetically modified maize MON810 and rice LLRICE62. Food Anal Method 6(2):705–713
Sun SL, Zhou YS, Chen J, Shi JP, Zhao HM, Hao HN, Song WB, Zhang M, Cui Y, Dong XM, Liu N, Ma XX, Jiao YP, Wang B, Wei XH, Stein JC, Glaubitz JC, Lu F, Yu GL, Fengler K, Li BL, Rafalski A, Schnable PS, Ware DH, Buckler ES, Lai JS (2018) Extensive intraspecific gene order and gene structural variations between Mo17 and other maize genomes. Na Genet 50:1289–1295
Wahler D, Schauser L, Bendiek J, Grohmann L (2013) Next-generation sequencing as a tool for detailed molecular characterisation of genomic insertions and flanking regions in genetically modified plants: a pilot study using a rice event unauthorised in the EU. Food Anal Methods 6(6):1718–1727
Wang J, Wang W, Li YR, Tian G, Goodman L, Fan W, Zhang JQ, Li J, Zhang JB, Guo YR, Feng B, Li H, Lu Y, Fang X, Liang H, Du Z, Li D, Zhao Y, Hu Y, Yang Z, Zheng H, Hellmann I, Inouye M, Pool J, Yi X, Zhao J, Duan J, Zhou Y, Qin J, Ma L, Li G, Yang Z, Zhang G, Yang B, Yu C, Liang F, Li W, Li S, Li D, Ni P, Ruan J, Li Q, Zhu H, Liu D, Lu Z, Li N, Guo G, Zhang J, Ye J, Fang L, Hao Q, Chen Q, Liang Y, Su Y, San A, Ping C, Yang S, Chen F, Li L, Zhou K, Zheng H, Ren Y, Yang L, Gao Y, Yang G, Li Z, Feng X, Kristiansen K, Wong GK, Nielsen R, Durbin R, Bolund L, Zhang X, Li S, Yang H, Wang J (2008) The diploid genome sequence of an Asian individual. Nature 456(7218):60–65
Wang M, Liu XX, Chang G, Chen YD, An G, Yan LY, Gao S, Xu YW, Cui YL, Dong J, Chen YH, Fan XY, Hu YQ, Song K, Zhu XH, Gao Y, Yao ZK, Bian SH, Hou Y, Lu JH, Wang R, Fan Y, Lian Y, Tang WH, Wang YP, Liu JQ, Zhan LM, Wang LY, Lou ZT, Yuan RP, Shi YJ, Hu BQ, Tang FC, Zhao XY, Qiao J (2019) Single-Cell RNA sequencing analysis reveals sequential cell fate transition during human spermatogenesis. Cell Stem Cell 23(4):599–614
Williams-Carrier R, Stiffler N, Belcher S, Kroeger T, Stem DB, Monde RA, Coalter R, Barkan A (2010) Use of Illumina sequencing to identify transposon insertions underlying mutant phenotypes in high-copy mutator lines of maize. Plant J 63(1):167
Wilson AK, Latham JR, Steinbrecher RA (2006) Transformation-induced mutations in transgenic plants: Analysis and biosafety implications. Biotechnol Genet Eng Rev 23(1):209–238
Windels P, Sylvie DB, Erik VB, De Loose M, Depicker A (2003) T-DNA integration in Arabidopsis chromosomes. Presence and origin of filler DNA sequences. Plant Physiol 133(4):2061–2068
Yang LT, Wang CM, Jensen AH, Morisset D, Lin YJ, Zhang DB (2013) Characterization of GM events by insert knowledge adapted re-sequencing approaches. Sci. Rep. 3:2839
Yang XD, Niu L, Zhang W, He HL, Yang J, Xing GJ, Guo DQ, Du Q, Qian XY, Yao Y, Li QY, Dong YS (2017) Robust RNAi-mediated resistance to infection of seven potyvirids in soybean expressing an intron hairpin NIb RNA. Transgenic Res 26(5):665–676
Acknowledgements
This work was supported by grants from China National Novel Transgenic Organisms Breeding Project (2016ZX08004-004). We would also like to thank Editage (www.editage.cn) for English language editing.
Author information
Authors and Affiliations
Contributions
XZ and XY designed the experiments and drafted the manuscript. LN and HH conducted the experiments and drafted the manuscript. Yuanyu Zhang analyzed the positions of inserted DNA. JY, GX, and HL grown soybean and other plant samples and collected them. All authors participated in manuscript revision.
Corresponding authors
Ethics declarations
Conflict of interest
The authors claim no conflict of interest in the publication of this paper.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Niu, L., He, H., Zhang, Y. et al. Efficient identification of genomic insertions and flanking regions through whole-genome sequencing in three transgenic soybean events. Transgenic Res 30, 1–9 (2021). https://doi.org/10.1007/s11248-020-00225-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11248-020-00225-8