Abstract
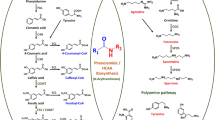
Rosmarinic acid (RA) and its derivatives have wide medicinal applications. However, the biosynthetic pathway of RA in Prunella vulgaris is still unclear. Here, we isolated and characterized a hydroxyphenylpyruvate reductase cDNA (PvHPPR, KM053279) from P. vulgaris, commonly known as “all-heal”. Based on the high sequence similarity of the predicted protein, PvHPPR was found to belong to the D-isomer-specific 2-hydroxyacid dehydrogenase family. Heterologous expression of the PvHPPR open reading frame was performed in Escherichia coli, and the resulting protein was proved to catalyze the NAD(P)H-dependent reduction of 4-hydroxyphenylpyruvate, phenylpyruvate, and pyruvate to the corresponding lactates. The Km values for the various substrates were 0.31 mM for 4-hydroxyphenylpyruvate, 0.73 mM for phenylpyruvate, and 1.02 mM for pyruvate, demonstrating that 4-hydroxyphenylpyruvate is the preferred substrate. The role of PvHPPR in RA biosynthesis was evaluated by using Agrobacterium rhizogenes-mediated overexpression in P. vulgaris hairy roots. PvHPPR transcript levels and RA contents were increased by 0.7–11.8-fold and 24–63% respectively compared with the wild-type counterparts, suggesting PvHPPR is likely involved in RA biosynthesis in P. vulgaris. The positive correlation between the two provides evidence for this claim.
Key message
A hydroxyphenylpyruvate reductase (HPPR) gene involved in rosmarinic acid biosynthesis in Prunella vulgaris was cloned, heterologously expressed, and functionally characterized.





Similar content being viewed by others
Data availability
All data generated or analyzed during this study are included in this published article [and its supplementary information fles].
Abbreviations
- HPPR:
-
Hydroxyphenylpyruvate reductase
- IPTG:
-
Isopropyl-b-D-thiogalactoside
- SDS:
-
Sodium dodecyl sulfate
- DTT:
-
Dithiothreitol
- RA:
-
Rosmarinic acid
- GFP:
-
Green fluorescence protein
- DW:
-
Dry weight
- HPP:
-
4-Hydroxyphenylpyruvate
References
Ahmad Z, Husaini A, Roslan HA (2015) Characterisation and expression analysis of hydroxyphenylpyruvate reductase derived from Orthosiphon aristatus. Borneo J Resour Sci Technol 5:34–42
Barberini S, Savona M, Raffi D, Leonardi M, La P, Stochmal A, Vainstein A, Pistelli Lu, Ruffoni B (2013) Molecular cloning of SoHPPR encoding a hydroxyphenylpyruvate reductase, and its expression in cell suspension cultures of Salvia officinalis. plant cell. Tissue Organ C (PCTOC) 114:131–138. https://doi.org/10.1007/s11240-013-0300-8
Bernard N, Johnsen K, Ferain T, Garmyn D, Hols P, Holbrook JJ, Delcour J (1994) NAD+-Dependent d-2-Hydroxyisocaproate dehydrogenase of Lactobacillus Delbrueckii subsp. Bulgaricus Eur J Biochem 224:439–446
Booth MP, Conners R, Rumsby G, Brady RL (2006) Structural basis of substrate specificity in human glyoxylate reductase/hydroxypyruvate reductase. J Mol Biol 360:178–189
Dengler U, Niefind K, Kieβ M, Schomburg D (1997) Crystal structure of a ternary complex of D-2-hydroxyisocaproate dehydrogenase from Lactobacillus casei, NAD+ and 2-oxoisocaproate at 1.9 Å resolution. J Mol Biol 267:640–660
Fujii T, Shimizu M, Doi Y, Fujita T, Ito T, Miura D, Wariishi H, Takaya N (2011) Novel fungal phenylpyruvate reductase belongs to d-isomer-specific 2-hydroxyacid dehydrogenase family. Biochimica Et Biophysica Acta (BBA) Proteins Proteomics 1814(12):1669–1676. https://doi.org/10.1016/j.bbapap.2011.05.024
Goldberg JD, Yoshida T, Brick P (1994) Crystal structure of a NAD-dependent d-glycerate dehydrogenase at 2· 4 Å resolution. J Mol Biol 236:1123–1140
Grant GA (1989) A new family of 2-hydroxyacid dehydrogenases. Biochem Biophys Res Commun 165:1371–1374
Häusler E, Petersen M, Alfermann AW (1991) Hydroxyphenylpyruvate reductase from cell suspension cultures of Coleus blumei Benth. Zeitschrift Für Naturforschung C 46:371–376
Hücherig S, Petersen M (2013) RNAi suppression and overexpression studies of hydroxyphenylpyruvate reductase (HPPR) and rosmarinic acid synthase (RAS) genes related to rosmarinic acid biosynthesis in hairy root cultures of Coleus blumei. Plant Cell. Tissue Organ Cult (PCTOC) 113:375–385. https://doi.org/10.1007/s11240-012-0277-8
Janiak V, Petersen M, Zentgraf M, Klebe G, Heine A (2010) Structure and substrate docking of a hydroxy (phenyl) pyruvate reductase from the higher plant Coleus blumei Benth. Acta Crystallogr D Biol Crystallogr 66:593–603
Kim KH, Janiak V, Petersen M (2004) Purification, cloning and functional expression of hydroxyphenylpyruvate reductase involved in rosmarinic acid biosynthesis in cell cultures of Coleus Blumei. Plant Mol Biol 54:311–323. https://doi.org/10.1023/B:PLAN.0000036367.03056.b2
Kim Y, Uddina M, Kim Y, Park C, Park S (2014) Molecular cloning and characterization of tyrosine aminotransferase and hydroxyphenylpyruvate reductase, and rosmarinic acid accumulation in Scutellaria baicalensis. Nat Prod Commun 9:1311–1314
Kumar S, Stecher G, Tamura K (2016) MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33:1870–1874. https://doi.org/10.1093/molbev/msw054
Kwon DY, Li X, Kim JK, Park SU (2019) Molecular cloning and characterization of rosmarinic acid biosynthetic genes and rosmarinic acid accumulation in Ocimum basilicum L. Saudi J Biol Sci 26:469–472. https://doi.org/10.1016/j.sjbs.2017.03.010
Lamzin VS, Dauter Z, Popov VO, Harutyunyan EH, Wilson KS (1994) High resolution structures of holo and apo formate dehydrogenase. J Mol Biol 236:759–785
Litvinenko V, Popova T, Simonjan A, Zoz I, Sokolov V (1975) “Tannins” and derivatives of hydroxycinnamic acid in labiatae. Planta Med 27:372–380
Mansouri M, Mohammadi F (2021) Transcriptome analysis to identify key genes involved in terpenoid and rosmarinic acid biosynthesis in lemon balm (Melissa officinalis). Gene 773:145417. https://doi.org/10.1016/j.gene.2021.145417
Nicholas K, Nicholas HJ (1997) GeneDoc: a tool for editing and annotating multiple sequence alignments. Embnew News 4:1–4
Osakabe N, Takano H, Sanbongi C, Yasuda A, Yanagisawa R, Inoue KI, Yoshikawa T (2004) Anti-inflammatory and anti-allergic effect of rosmarinic acid (RA); inhibition of seasonal allergic rhinoconjunctivitis (SAR) and its mechanism. BioFactors 21:127–131
Parnham M, Kesselring K (1985) Rosmarinic acid. Drugs Future 10:756–757
Petersen M (1997) Cytochrome P450-dependent hydroxylation in the biosynthesis of rosmarinic acid in Coleus. Phytochemistry 45:1165–1172
Petersen M, Alfermann A (1988) Two new enzymes of rosmarinic acid biosynthesis from cell cultures of Coleus blumei: hydroxyphenylpyruvate reductase and rosmarinic acid synthase. Zeitschrift Für Naturforschung C 43:501–504
Petersen M, Simmonds MS (2003) Rosmarinic acid. Phytochemistry 62:121–125
Petersen M, Häusler E, Karwatzki B, Meinhard J (1993) Proposed biosynthetic pathway for rosmarinic acid in cell cultures of Coleus blumei Benth. Planta 189:10–14
Razeto A, Kochhar S, Hottinger H, Dauter M, Wilson KS, Lamzin VS (2002) Domain closure, substrate specificity and catalysis of D-lactate dehydrogenase from Lactobacillus bulgaricus. J Mol Biol 318:109–119
Ru M, An Y, Wang K, Peng L, Li B, Bai Z, Wang B, Liang Z (2016) Prunella vulgaris L. hairy roots: Culture, growth, and elicitation by ethephon and salicylic acid. Eng Life Sci 16:494–502. https://doi.org/10.1002/elsc.201600001
Ru M, Wang K, Bai Z, Peng L, He S, Pei T, Jia Y, Li H, Liang Z (2017a) Molecular cloning and characterisation of two enzymes involved in the rosmarinic acid biosynthesis pathway of Prunella vulgaris L. plant Cell. Tissue Organ Cult (PCTOC) 128:381–390. https://doi.org/10.1007/s11240-016-1117-z
Ru M, Wang K, Bai Z, Peng L, He S, Wang Y, Liang Z (2017b) A tyrosine aminotransferase involved in rosmarinic acid biosynthesis in Prunella vulgaris L. Sci Rep 7:4892. https://doi.org/10.1038/s41598-017-05290-4
Scarpati M, Oriente G (1958) Isolamento e costituzione dell’acido rosmarinico (dal rosmarinus off.). Riserca Sci 28:2329–2333
Schuller DJ, Grant GA, Banaszak LJ (1995) The allosteric ligand site in the Vmax-type cooperative enzyme phosphoglycerate dehydrogenase. Nat Struct Mol Biol 2:69–76
Tanaka N, Nonaka T, Nakanishi M, Deyashiki Y, Hara A, Mitsui Y (1996) Crystal structure of the ternary complex of mouse lung carbonyl reductase at 1.8 A resolution: the structural origin of coenzyme specificity in the short-chain dehydrogenase/reductase family. Structure 4:33–45
Timm S, Nunes-Nesi A, Pärnik T, Morgenthal K, Wienkoop S, Keerberg O, Weckwerth W, Kleczkowski LA, Fernie AR, Bauwe H (2008) A cytosolic pathway for the conversion of hydroxypyruvate to glycerate during photorespiration in Arabidopsis. Plant Cell 20:2848–2859
Wang G, Chen J, Yi B, Tan H, Zhang L, Chen W (2017) HPPR encodes the hydroxyphenylpyruvate reductase required for the biosynthesis of hydrophilic phenolic acids in Salvia miltiorrhiza. Chin J Nat Med 15:917–927
Xiao Y, Zhang L, Gao S, Saechao S, Di P, Chen J, Chen W (2011) The c4h, tat, hppr and hppd genes prompted engineering of rosmarinic acid biosynthetic pathway in Salvia miltiorrhiza hairy root cultures. PLoS ONE 6:e29713. https://doi.org/10.1371/journal.pone.0029713
Xiao Y (2009) Study on biosynthetic regulation of phenolic acids in Salvia miltiorrhiza Bunge. Doctoral Dissertation, Shanghai: Second Military Medical University
Acknowledgements
This work was financially supported by the Science & Technology and Special Talents Project of Guangxi (Guike AD19245087) and the National Natural Science Foundation of China (Grant no: 81373908, 82160754).
Funding
National Natural Science Foundation of China, 81373908, Zongsuo Liang, Science & Technology and Special Talents Project of Guangxi, Guike AD19245087,Mei Ru
Author information
Authors and Affiliations
Contributions
RM and LZS: conceived and designed research. RM, WCM and LP: conducted experiments. RM, CLY, and LJL: analyzed data. RM, LTT, and LZS: wrote the manuscript. RM, YCC, TXY, and LZS: revised the manuscript. All authors have read and approved the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors have no conflict of interest to declare.
Additional information
Communicated by Ali R. Alan.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Ru, M., Chen, L., Liu, J. et al. Cloning, heterologous expression, and functional characterization of a hydroxyphenylpyruvate reductase (HPPR) gene involved in rosmarinic acid biosynthesis in Prunella vulgaris. Plant Cell Tiss Organ Cult 153, 273–283 (2023). https://doi.org/10.1007/s11240-023-02452-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11240-023-02452-3