Abstract
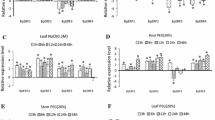
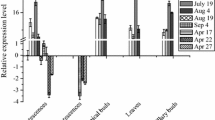
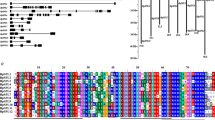
TEOSINTE-LIKE1, CYCLOIDEA, and PROLIFERATING CELL FACTOR1 (TCP) genes are plant-specific transcription factors, which play important roles in plant growth and development. The function of CIN subclass of the TCP gene family has not been well revealed. In this investigation, five CIN subfamily members were identified in Betula platyphylla, all of which contained the conservative domains associated with the CIN subclass. Most of the genes were involved in the development of leaves, apical buds, and male inflorescences, especially the BpTCP7 gene. The results of GUS activity assays driven by BpTCP7 promoters confirmed these conclusions. The analysis of the BpTCP7 promoter also showed that the gene was induced by salt and drought stresses, and multiple hormones. Through Agrobacterium-mediated genetic transformation, we obtained four independent birch lines with over-expression of BpTCP7. Moreover, physiological and enzymatic analysis of transgenic lines under salt and drought stress showed that the scavenging effect of reactive oxygen species (ROS) of transgenic lines was improved. Our results provide useful information for revealing the functions of CIN subclasses in the growth and development of B. platyphylla and their applications in scavenging of ROS.
Key message
Overexpression of BpTCP7 gene leaded to improved scavenging reactive oxygen species (ROS) in drought and salinity treatment through multiple hormone pathways in Betula platyphylla.









Similar content being viewed by others
Abbreviations
- TCP:
-
TEOSINTE-LIKE1, CYCLOIDEA, and PROLIFERATING CELL FACTOR1
- CIN:
-
CINCINNATA
- ROS:
-
Reactive oxygen species
- qRT-PCR:
-
Quantificational real-time polymerase chain
- SOD:
-
Superoxide dismutase
- ORF:
-
Open reading frame
- WPM:
-
Woody plant medium
References
Aguilar-Martinez JA, Poza-Carrion C, Cubas P (2007) Arabidopsis BRANCHED1 acts as an integrator of branching signals with in axillary buds. Plant Cell 19:458–472
Almeida DM, Gregorio GB, Oliveira MM, Saibo NJ (2017) Five novel transcription factors as potential regulators of OsNHX1 gene expression in a salt tolerant rice genotype. Plant Mol Biol 93(1–2):61–77
Arnon DI (1949) Copper enzymes in isolated chloroplasts, polyphenoloxidase in Beta vulgaris. Plant Physiol 24:1–15
Chen L, Jiang BJ, Wu CX, Sun S, Hou WS, Han TF (2015) The characterization of GmTIP, a root-specific gene from soybean, and the expression analysis of its promoter. Plant Cell Tissue Organ Cult 121:259–274
Classic Murashige T, Skoog F (1962) A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol Plant 15:473–497
Clough SJ, Bent AF (1998) Floral dip: a simplified method for agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16:735–743
Cubas P, Lauter N, Doebley J, Coen E (1999) The TCP domain: a motif found in proteins regulating plant growth and development. Plant J 18:215–222
Danisman S, van der Wal F, Dhondt S, Waites R, de Folter S, Bimbo A, van Dijk AD, Muino JM, Cutri L, Dornelas MC, Angenent GC, Immink RG (2012) Arabidopsis class I and class II TCP transcription factors regulate jasmonic acid metabolism and leaf development antagonistically. Plant Physiol 159(4):1511–1523
Doebley J, Stec A, Hubbard L (1997) The evolution of apical dominance in maize. Nature 386(6624):485–488
Efroni I, Blum E, Goldshmidt A, Eshed Y (2008) A protracted and dynamic maturation schedule underlies Arabidopsis leaf development. Plant Cell 20(9):2293–2306
Francis A, Dhaka N, Bakshi M, Jung KH, Sharma MK, Sharma R (2016) Comparative phylogenomic analysis provides insights into TCP gene functions in Sorghum. Sci Rep 6:38488
Higo K, Ugawa Y, Iwamoto M, Korenaga T (1999) Plant cis-acting regulatory DNA elements (PLACE) database: 1999. Nucleic Acids Res 27:297–300
Jefferson RA, Kavanagh TA, Bevan MW (1987) GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J 6(13):3901–3907
Kosugi S, Ohashi Y (2002) DNA binding and dimerization specificity and potential targets for the TCP protein family. Plant J 30(3):337–348
Koyama T, Furutani M, Tasaka M, Ohme-Takagi M (2007) TCP transcription factors control the morphology of shoot lateral organs via negative regulation of the expression of boundary-specific genes in Arabidopsis. Plant Cell 19(2):473–484
Koyama T, Mitsuda N, Seki M, Shinozaki K, Ohme-Takagi M (2010) TCP transcription factors regulate the activities of ASYMMETRIC LEAVES1 and miR164, as well as the auxin response, during differentiation of leaves in Arabidopsis. Plant Cell 22:3574–3588
Kubota A, Ito S, Shim JS, Johnson RS, Song YH, Breton G, Goralogia GS, Kwon MS, Laboy Cintrón D, Koyama T, Ohme-Takagi M, Pruneda-Paz JL, Kay SA, MacCoss MJ, Imaizumi T (2017) TCP4-dependent induction of CONSTANS transcription requires GIGANTEA in photoperiodic flowering in Arabidopsis. PLoS Genet 13(6):e1006856
Lescot M, Dehais P, Thijs G, Marchal K, Moreau Y, Van de Peer Y, Rouzé P, Rombauts S (2002) PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res 30:325–327
Li S, Zachgo S (2013) TCP3 interacts with R2R3-MYB proteins,promotes flavonoid biosynthesis and negatively regulates the auxin response in Arabidopsis thaliana. Plant J 76(6):901–913
Li HY, Jiang J, Wang S, Liu FF (2010a) Expression analysis of ThGLP, a new germin-like protein gene, in Tamarix hispida. J For Res 21(3):323–330
Li QF, Sun SM, Yuan DY, Yu HX, Gu MH, Liu QQ (2010b) Validation of candidate reference genes for the accurate normalization of real-time quantitative RT-PCR data in rice during seed development. Plant Mol Biol Rep 28:49–57
Li Z, Li B, Shen WH, Huang H, Dong A (2012) TCP transcription factors interact with AS2 in the repression of class-I KNOX genes in Arabidopsis thaliana. Plant J 71(1):99–107
Liu BX, Wang ZG, Liang HY, Yang MS (2012) Effects of salt stress on physiological characters and salt-tolerance of Ulmus pumila in different habitats. Chin J Appl Ecol 23(6):1481–1489
Martin-Trillo M, Cubas P (2010) TCP genes: a family snapshot ten years later. Trends Plant Sci 15:31–39
Masuda HP, Cabral LM, De Veylder L, Tanurdzic M, de Almeida Engler J, Geelen D, Inzé D, Martienssen RA, Ferreira PC, Hemerly AS (2008) ABAP1 is a novel plant Armadillo BTB protein involved in DNA replication and transcription. EMBO J 27(20):2746–2756
Mukhopadhyay P, Tyagi AK (2015) OsTCP19 influences developmental and abiotic stress signaling by modulating ABI4-mediated pathways. Sci Rep 5:9998
Nath U, Aggarwal P, Challa KR (2016) Activation of YUCCA5 by the transcription factor TCP4 integrates developmental and environmental signals to promote hypocotyl elongation in Arabidopsis. Plant Cell 28:2117–2130
Navaud O, Dabos P, Carnus E, Tremousaygue D, Hervé C (2007) TCP transcription factors predate the emergence of land plants. J Mol Evol 65(1):23–33
Palatnik JF, Allen E, Wu X, Schommer C, Schwab R, Carrington JC, Weigel D (2003) Control of leaf morphogenesis by microRNAs. Nature 425(6955):257–263
Parapunova V, Busscher M, Busscher-Lange J, Lammers M, Karlova R, Bovy AG (2014) Identification, cloning and characterization of the tomato TCP transcription factor family. BMC Plant Biol 14:157
Parida AK, Das AB (2005) Salt tolerance and salinity effects on plants. Ecotoxicol Environ Safe 60:324–349
Sarvepalli K, Nath U (2011) Hyper-activation of the TCP4 transcription factor in Arabidopsis thaliana accelerates multiple aspects of plant maturation. Plant J 67:595–607
Schmittgen TD, Livak KJ (2008) Analyzing real-time PCR data by the comparative C T method. Nat Protoc 3(6):1101–1108
Schommer C, Palatnik JF, Aggarwal P, Chételat A, Cubas P, Farmer EE, Nath U, Weigel D (2008) Control of jasmonate biosynthesis and senescence by miR319 targets. PLoS Biol 6(9):e230
Shleizer-Burko S, Burko Y, Ben-Herzel O, Ori N (2011) Dynamic growth program regulated by LANCEOLATE enables flexible leaf patterning. Development 138(4):695–704
Tao J, Zhan YG, Jiang J, Yang CP, Liu YX (1998) Tissue culture and regenerative system of Betula platyphylla Suks. J Northeast For Univ 26(5):6–9
Tao Q, Guo D, Wei B, Zhang F, Pang C, Jiang H, Zhang J, Wei T, Gu H, Qu LJ, Qin G (2013) The TIE1 transcriptional repressor links TCP transcription factors with TOPLESS/TOPLESS-RELATED corepressors and modulates leaf development in Arabidopsis. Plant Cell 25(2):421–437
Vadde BVL, Challa KR, Nath U (2018) The TCP4 transcription factor regulates trichome cell differentiation by directly activating GLABROUS INFLORESCENCE STEMS in Arabidopsis thaliana. Plant J 93(2):259–269
Wang DM, Jia Y, Cui JZ (2009) Advances in research on effects of salt stress on plant and adaptive mechanism of the plant to salinity. Chin Agric Sci Bull 25(04):124–128
Wang ST, Sun XL, Hoshino Y, Yu Y, Jia B, Sun ZW, Sun MZ, Duan XB, Zhu YM (2014a) MicroRNA319 positively regulates cold tolerance by targeting OsPCF and OsTCP21 in rice (Oryza sativa L.). PLoS ONE 9(3):e91357
Wang X, Jiang Q, Wang W, Su L, Han Y, Wang C (2014b) Molecular mechanism of polypeptides from Chlamys farreri (PCF)'s anti-apoptotic effect in UVA-exposed HaCaT cells involves HSF1/HSP70, JNK, XO, iNOS and NO/ROS. J Photochem Photobiol B 130:47–56
Xu R, Sun P, Jia F, Lu L, Li Y, Zhang S (2014) Genome wide analysis of TCP transcription factor gene family in Malus domestica. Genetics 93:733–746
Yang G, Chen S, Wang S, Liu GF, Li HY, Huang HJ, Jiang J (2015) BpGH3.5, an early auxin-response gene, regulates root elongation in Betula platyphylla × Betula pendula. Plant Cell Tissue Org 120(1):239–250
Zhan YG, Liu ZH, Wang YC, Wang ZY, Yang CP, Liu GF (2001) Transformation of insect resistant gene into birch. J Northeast For Univ 29(6):4–6
Zhou M, Li D, Li Z, Hu Q, Yang C, Zhu L, Luo H (2013) Constitutive expression of a miR319 gene alters plant development and enhances salt and drought tolerance in transgenic creeping bentgrass. Plant Physiol 161(3):1375–1391
Funding
This work was supported by the Fundamental Research Funds for the Central Universities (No. 2572018BW03), the Innovation Project of State Key Laboratory of Tree Genetics and Breeding (Northeast Forestry University) (2015B01), the 111 Project (B16010), Provincial Funding for Major National Science and Technology Projects and Key R&D Projects in Heilongjiang Province (GX18B027).
Author information
Authors and Affiliations
Contributions
JJ designed and managed the research work and improved the manuscript; HL designed the research work and wrote the manuscript; HY wrote the manuscript; LA performed the bioinformatics analysis; FM, YY and JL performed the experiments; LR prepared all figures. All the authors reviewed the manuscript.
Corresponding author
Additional information
Communicated by Sergio J. Ochatt.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Li, H., Yuan, H., Liu, F. et al. BpTCP7 gene from Betula platyphylla regulates tolerance to salt and drought stress through multiple hormone pathways. Plant Cell Tiss Organ Cult 141, 17–30 (2020). https://doi.org/10.1007/s11240-019-01748-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11240-019-01748-7