Abstract
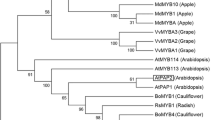
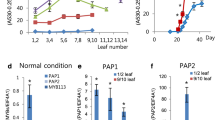
The R2R3-Myb proteins comprise a large family of plant transcription factors that regulate the expression of multiple drought-responsive and cold-responsive genes by binding special cis-elements at the promoter. There have been previous studies in transgenic plant over-expression of the myb protein. In this article, we analyzed the function of MdMYB6, an R2R3-type Myb transcription factor from apples (Malus × domestica), in transgenic Arabidopsis plants. In contrast to wild type plants, these transgenic lines accumulated less anthocyanin. Equimolar concentrations of sorbitol did not alter the anthocyanin accumulation. Real-time polymerase chain reaction (PCR) indicated that MdMYB6 over-expression suppressed enzymes involved in anthocyanin biosynthesis and some Arabidopsis basic helix-loop-helix (bHLH) genes. The MdMYB6 transcription faction may be an important repressor of anthocyanin biosynthesis in plants.




Similar content being viewed by others
References
Baier M, Hemmann G, Holman R, Corke F, Card R, Smith C, Rook F, Bevan MW (2004) Characterization of mutants in Arabidopsis showing increased sugar-specific gene expression, growth, and developmental responses. Plant Physiol 134:81–91
Ban Y, Honda C, Hatsuyama Y, Igarashi M, Bessho H, Moriguchi T (2007) Isolation and functional analysis of a MYB transcription factor gene that is a key regulator for the development of red coloration in apple skin. Plant Cell Physiol 48(7):958–970
Baud S, Vaultier MN, Rochat C (2004) Structure and expression profile of the sucrose synthase multigene family in Arabidopsis. J Exp Bot 55:397–409
Borevitz JO, Xia Y, Blount J, Dixon RA, Lamb C (2000) Activation tagging identifies a conserved MYB regulator of phenylpropanoid biosynthesis. Plant Cell 12:2383–2394
Broun P (2004) Transcription factors as tools for metabolic engineering in plants. Curr Opin Plant Biol 7:202–209
Carey CC, Strahle JT, Selinger DA, Chandler VL (2004) Mutations in the pale aleurone color1 regulatory gene of the Zea mays anthocyanin pathway have distinct phenotypes relative to the functionally similar TRANSPARENT TESTA GLABRA1 gene in Arabidopsis thaliana. Plant Cell 16:450–464
Chandran D, Reinders A, Ward JM (2003) Substrate specificity of the Arabidopsis thaliana sucrose transporter AtSUC2. J Biol Chem 278:44320–44325
Chen YH, Yang XY, He K, Liu MH, Li JG, Gao ZF, Lin ZQ, Zhang YF, Wang XX, Qiu XM, Shen YP, Zhang L, Deng XH, Luo JC, Deng XW, Chen ZL, Gu HY, Qu L-J (2006) The MYB transcription factor superfamily of Arabidopsis: expression analysis and phylogenetic comparison with the rice MYB family. Plant Mol Biol 60(1):107–124
Déjardin A, Sokolov LN, Kleczkowski LA (1999) Sugar/osmoticum levels modulate differential abscisic acid-independent expression of two stress-responsive sucrose synthase genes in Arabidopsis. Biochem J 344(Pt 2):503–509
Espley RV, Hellens RP, Putterill J, Stevenson DE, Kutty-Amma S, Allan AC (2007) Red colouration in apple fruit is due to the activity of the MYB transcription factor, MdMYB10. Plant J. 49:414–427
Gollop R, Even S, Colova-Tsolova V, Perl A (2002) Expression of the grape dihydroflavonol reductase gene and analysis of its promoter region. J Exp Bot 53:1397–1409
Gonzalez A, Zhao M, Leavitt JM, Lloyd AM (2008) Regulation of the anthocyanin biosynthetic pathway by the TTG1/bHLH/Myb transcriptional complex in Arabidopsis seedlings. Plant J 53(5):814–827
Han YP, Vimolmangkang S, Soria-Guerra RE, Rosales-Mendoza S, Zheng D, Lygin AV, Korban SS (2010) Ectopic expression of apple F3′H genes contributes to anthocyanin accumulation in the Arabidopsis tt7 mutant grown under nitrogen stress. Plant Physiol 153(2):806–820
Hellens RP, Allan AC, Friel EN, Bolitho K, Grafton K, Templeton MD, Karunairetnam S, Gleave AP, Laing WA (2005) Transient expression vectors for functional genomics, quantification of promoter activity and RNA silencing in plants. Plant Methods 1:13
Holton TA, Cornish EC (1995) Genetics and biochemistry of anthocyanin biosynthesis. Plant Cell 7:1071–1083
Huijser C, Kortstee A, Pego J, Weisbeek P, Wisman E, Smeekens S (2000) The Arabidopsis SUCROSE UNCOUPLED-6 gene is identical to ABSCISIC ACID INSENSITIVE-4: involvement of abscisic acid in sugar responses. Plant J 23:577–585
Jang JC, León P, Zhou L, Sheen J (1997) Hexokinase as a sugar sensor in higher plants. Plant Cell 9:5–19
Jin H, Cominelli E, Bailey P, Parr A, Mehrtens F, Jones J, Tonelli C, Weisshaar B, Martin C (2000) Transcriptional repression by AtMYB4 controls production of UV-protecting sunscreens in Arabidopsis. EMBO J 19:6150–6161
Koes R, Verweij W, Quattrocchio F (2005) Flavonoids: a colorful model for the regulation and evolution of biochemical pathways. Trends Plant Sci 10:236–242
Martin C, Paz-Ares J (1997) MYB transcription factors in plants. Trends Genet 13(2):67–73
Matsui K, Tanaka H, Ohme-Takagi M (2004) Suppression of the biosynthesis of proanthocyanidin in Arabidopsis by a chimeric PAP1 repressor. Plant Biotechnol J 2:487–493
Meyer S, Lauterbach C, Niedermeier M, Barth I, Sjolund RD, Sauer N (2004) Wounding enhances expression of AtSUC3, a sucrose transporter from Arabidopsis sieve elements and sink tissues. Plant Physiol 134:684–693
Mita S, Hirano H, Nakamura K (1997) Negative regulation in the expression of a sugar-inducible gene in Arabidopsis thaliana: a recessive mutation causing enhanced expression of a gene for beta-amylase. Plant Physiol 114:575–582
Nesi N, Debeaujon I, Jond C, Pelletier G, Caboche M, Lepiniec L (2000) The TT8 gene encodes a basic helix-loop-helix domain protein required for expression of DFR and BAN genes in Arabidopsis siliques. Plant Cell 12:1863–1878
Ohto M, Onai K, Furukawa Y, Aoki E, Araki T, Nakamura K (2001) Effects of sugar on vegetative development and floral transition in Arabidopsis. Plant Physiol 127:252–261
Park KL, Ishikawa N, Morita Y, Choi JD, Hoshino A, Iida S (2007) A bHLH regulatory gene in the common morning glory, Ipomoea purpurea, controls anthocyanin biosynthesis in flowers, proanthocyanidin and phytomelanin pigmentation in seeds, and seed trichome formation. Plant J 49(4):641–654
Ramsay NA, Glover BJ (2005) MYB-bHLH-WD40 protein complex and the evolution of cellular diversity. Trends Plant Sci 10:63–70
Schulze W, Weise A, Frommer WB, Ward JM (2000) Function of the cytosolic N-terminus of sucrose transporter AtSUT2 in substrate affinity. FEBS Lett 485:189–194
Shirley BW, Kubasek WL, Storz G, Bruggemann E, Koornneef M, Ausubel FM, Goodman HM (1995) Analysis of Arabidopsis mutants deficient in flavonoid biosynthesis. Plant J 8:659–671
Sivitz AB, Reinders A, Ward JM (2008) Arabidopsis sucrose transporter AtSUC1 is important for pollen germination and sucrose-induced anthocyanin accumulation. Plant Physiol 147:92–100
Teng S, Keurentjes J, Bentsink L, Koornneef M, Smeekens S (2005) Sucrose-specific induction of anthocyanin biosynthesis in Arabidopsis requires the MYB75/PAP1 gene. Plant Physiol 139:1840–1852
Tsukaya H, Ohshima T, Naito S, Chino M, Komeda Y (1991) Sugar-dependent expression of the CHS-A gene for chalcone synthase from petunia in transgenic Arabidopsis. Plant Physiol 97:1414–1421
Wade HK, Sohal AK, Jenkins GI (2003) Arabidopsis ICX1 is a negative regulator of several pathways regulating flavonoid biosynthesis genes. Plant Physiol 131:707–715
Ward JM, Kühn C, Tegeder M, Frommer WB (1998) Sucrose transport in higher plants. Int Rev Cytol 178:41–71
Winkel-Shirley B (2001) Flavonoid biosynthesis. A colorful model for genetics, biochemistry, cell biology, and biotechnology. Plant Physiol 126(2):485–493
Zhang X, Henriques R, Lin SS, Niu QW, Chua NH (2006) Agrobacterium-mediated transformation of Arabidopsis thaliana using the floral dip method. Nat Protoc 1:641–646
Zhu B, Xiong AS, Peng RH, Xu J, Zhou J, Xu JT, Jin XF, Zhang Y, Hou XL, Yao QH (2008) Heat stress protection in Aspen sp1 transgenic Arabidopsis thaliana. BMB Rep 41(5):382–387
Acknowledgments
This research was supported by the Shanghai National Natural Science Foundation (30670179, 30800602, 08ZR1417200); 863 Program (2006AA10Z117, 2006AA06Z358, 2008AA10Z401); Shanghai Project for ISTC (08540706500); The Key Project Fund of the Shanghai Municipal Committee of Agriculture (No. 2008-7-5); and The Shanghai Rising-Star Program (08QH14021).
Author information
Authors and Affiliations
Corresponding author
Additional information
J.-J. Gao and X.-F. Shen contributed equally to this article.
Rights and permissions
About this article
Cite this article
Gao, JJ., Shen, XF., Zhang, Z. et al. The myb transcription factor MdMYB6 suppresses anthocyanin biosynthesis in transgenic Arabidopsis. Plant Cell Tiss Organ Cult 106, 235–242 (2011). https://doi.org/10.1007/s11240-010-9912-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11240-010-9912-4