Abstract
Faecal samples (n = 1,093) collected from the woylie Bettongia penicillata Gray, in south-western Australia were examined for the presence of coccidian parasites. Eimeria sp. oöcysts were detected in 15.2% of samples. Faecal samples obtained from the eastern bettong Bettongia gaimardi (Desmarest) (n = 4) and long-nosed potoroo Potorous tridactylus (Kerr) (n = 12) in Tasmania, were also screened for the presence of Eimeria spp. (prevalence 50% and 41.7%, respectively). Morphological and genetic comparison with other known species of Eimeria indicates that the material identified in woylies is novel. This study aimed to (i) morphologically describe and genetically characterise Eimeria woyliei n. sp. found in woylies; and (ii) genetically characterise Eimeria gaimardi Barker, O’Callaghan & Beveridge, 1988, Eimeria potoroi Barker, O’Callaghan & Beveridge, 1988, and Eimeria mundayi Barker, O’Callaghan & Beveridge, 1988, from other potoroid marsupials. Molecular phylogenetic analyses conducted at the 18S rDNA and mitochondrial cytochrome c oxidase subunit 1 (cox1) loci revealed that E. woyliei n. sp. was most closely related to Eimeria setonicis Barker, O’Callaghan & Beveridge, 1988, at the 18S rDNA locus, and Eimeria trichosuri O’Callaghan & O’Donoghue, 2001, at the cox1 locus. Eimeria woyliei n. sp. is the sixth species of Eimeria to be formally described from potoroid marsupials.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Coccidian parasites are known to infect potoroid marsupials, including the critically endangered woylie or brush-tailed bettong Bettongia penicillata Gray. Although morbidity and mortality associated with coccidial infection in free-ranging macropods is uncommon (Vogelnest & Portas, 2008), disease has been documented in macropods under stress (e.g. the eastern grey kangaroo Macropus giganteus Shaw; Barker et al., 1972). Given the reliance of many threatened species on interventional management practices such as translocation, a process which has been identified as a significant stressor (Hing et al., 2017), it is imperative that we gain a greater understanding of the parasite species infecting wildlife, particularly those with the potential to cause disease in their host (e.g. coccidian parasites).
To date, no coccidian parasites (Apicomplexa: Eimeriidae) have been formally described in woylies (Duszynski, 2016), though Eimeria sp. oöcysts have been detected in faecal samples (Northover et al., 2017). Five Eimeria spp. including Eimeria potoroi Barker, O’Callaghan & Beveridge, 1988, Eimeria mundayi Barker, O’Callaghan & Beveridge, 1988, Eimeria gaimardi Barker, O’Callaghan & Beveridge, 1988, Eimeria aepyprymni Barker, O’Callaghan & Beveridge, 1988, and Eimeria burdi Hulst, Kemp & Slapeta, 2016, have been morphologically described from potoroid marsupials (Marsupialia: Potoroidae). Eimeria gaimardi from the eastern bettong Bettongia gaimardi (Desmarest), most closely morphologically resembles Eimeria sp. oöcysts found in woylies. Unfortunately, genetic characterisation of potoroid Eimeria spp. has not been undertaken. While Eimeria spp. tend to be host-specific (Barker et al., 1988), members of the genus Macropus Shaw have been known to harbor the same Eimeria spp. (e.g. Eimeria macropodis Wenyon & Scott, 1925; see Barker et al., 1989), thus it is important that additional data (e.g. genetic characterisation) is used to support the description of new species.
During this study we aimed to (i) morphologically describe and genetically characterise E. woyliei n. sp. from woylies; and (ii) genetically characterise E. gaimardi from the eastern bettong, and E. potoroi and E. mundayi from the long-nosed potoroo Potorous tridactylus (Kerr).
Materials and methods
Sample collection
Between 2014 and 2018, woylie faecal samples (n = 1,093) were collected from various sites within south-western Australia (Table 1) as part of a collaborative project with the Department of Biodiversity, Conservation and Attractions (DBCA), Kensington, Australia. Newspaper was placed beneath each trap to collect faeces, which were stored in 70% ethanol, 10% buffered formalin and/or 2% potassium dichromate until processing.
In 2018, faecal samples collected from the eastern bettong (n = 4) were obtained from a captive population at Bonorong Wildlife Sanctuary in Brighton, Tasmania (42.71°S, 147.27°E). Samples from the long-nosed potoroo (n = 12) were acquired from wild-caught animals within the Peter Murrell reserves, south of Hobart, Tasmania (43.00°S, 147.18°E). Faecal samples from the eastern bettong and long-nosed potoroo were collected directly from traps and samples were stored in 2% potassium dichromate prior to analysis.
Identification of coccidian oöcysts in faecal samples
For woylies, the majority of faecal samples (n = 1,073) were examined for the presence of coccidian oöcysts using simple faecal flotation with sodium nitrate (NaNO3) as described by Northover et al. (2017). These samples were formalin-fixed. To describe the morphology of E. woyliei n. sp., an additional 20 samples were sporulated (using potassium dichromate) and underwent faecal flotation using zinc sulphate (ZnS04) in distilled water (SG 1.20). Briefly, faeces were placed into a 10 ml centrifuge tube (up to the 1.5–2.0 ml mark), emulsified in distilled water (tube filled to the 10-ml mark) and centrifuged (2,000 rpm for 2 min). The supernatant was removed before filling the tube with zinc sulphate solution and re-emulsifying, before final centrifugation (2,000 rpm for 2 min). A sterile wire loop was used to transfer 2–3 loops from the surface of the tube to a glass slide, and a 22 × 22 mm coverslip was placed on top. Each sample was examined at 100× magnification using an Olympus BX50 microscope. All other potoroid samples were examined using zinc sulphate. To calculate the prevalence of infection, each faecal sample was scored as positive or negative for the presence of coccidian oöcysts.
Morphological description of the new species
Eighty-six sporulated oöcysts from a single woylie originating from Perup Sanctuary were examined at magnifications of 400–1,000×, using an Olympus BX50 microscope with a Olympus DP71 Universal Camera with Cellsens software. Photographs of sporulated oöcysts were taken using bright field microscopy. Measurements of oöcyst and sporocyst length and width, and oöcyst wall thickness were acquired using ImageJ software (US National Institute of Health, Bethesda, Maryland). All measurements are recorded in micrometres (µm) with the range followed by the mean in parentheses. We measured a single laterally positioned sporocyst within each oöcyst; if we could not identify a sporocyst in the correct position, we did not measure sporocyst length or width.
Morphological identification of other potoroid Eimeria spp.
Three distinct morphotypes of sporulated oöcysts were identified from the faeces of the eastern bettong and long-nosed potoroo. Based on their size and unique oöcyst and sporocyst characters as described by Barker et al. (1988) we identified E. gaimardi from the eastern bettong, and E. mundayi and E. potoroi from the long-nosed potoroo.
Genetic characterisation of Eimeria spp. in potoroid marsupials
For the woylie, nine faecal samples (eight ethanol- and one potassium dichromate-preserved) were used to genetically characterise E. woyliei n. sp. at the 18S rDNA and mitochondrial cytochrome c oxidase subunit 1 (cox1) loci. Eimeria gaimardi, E. mundayi and E. potoroi were genetically characterised using a single faecal sample (potassium dichromate-preserved) for each Eimeria species; as outlined above, morphological identification of sporulated oöcysts confirmed the identity of each species prior to genetic characterisation.
DNA extraction and PCR amplification
Samples stored in 70% ethanol and/or 2% potassium dichromate were exposed to four freeze/thaw cycles as described by Yang et al. (2016a) in order to achieve oöcyst lysis. Following the freeze/thaw step, faecal samples stored in 2% potassium dichromate were subjected to a wash step prior to lysis in order to remove the fixative, by centrifuging at 3000 rpm for 10 min and resuspending in phosphate buffered solution. DNA was isolated from 0.25 g of faecal sample using the PowerFecal DNA Isolation Kit (MolBio, Carlsbad, California) as per the manufacturer’s instructions. A negative control was included to rule out contamination.
Faecal samples from the woylie were screened at the 18S rDNA locus using a nested PCR with the external Eimeria spp. primers EiF1 (5′-GCT TGT CTC AAA GAT TAA GCC-3′) and EiR3 (5′-ATG CAT ACT CAA AAG ATT ACC-3′), and the internal Eimeria spp. primers EiR4 (5′-ACT CAA AAG ATT ACC TAG AC-3′) and EiF4 (5′-CTA TGG CTA ATA CAT GCG CAA T-3′) (Yang et al., 2016a). PCR reactions were carried out in a total volume of 25 µl containing 12.5 µl of 2× KAPA HiFi Hotstart ReadyMix (Millennium Science Pty. Ltd, Victoria, Australia), 0.75 µl primer (10 µM) and 2 µl DNA template. All PCR reactions were performed as described by Yang et al. (2016a) consisting of a pre-PCR step of 94°C for 3 min, followed by 45 cycles of 94°C for 30 s, 55°C annealing temperature for 30 s and 72°C for 2 min, and a final extension step of 72°C for 5 min.
Faeces from the eastern bettong and long-nosed potoroo were screened using the 18S external primers EiGTF1 (5′-TTC ACT GGT CCC TCC GAT C-3′) and EiGTR1 (5′-AAC CAT GGT AAT TCT ATG G-3′) (Yang et al., 2016b), and the internal primers EiGTF2 (5′-TTA CGC CTACTA GGC ATT CC-3′) and EiTR2 (5′-TGA CCT ATC AGC TTT CGA CG-3′). The PCR reaction contained 10 µl 2× GoTaq PCR master mix (Promega, Alexandria NSW, Australia), 1 µl DNA (50 ng), 10 µl of each primer (10 µM stock) and 7 µl distilled water. PCR cycling conditions for the external PCR were 1 cycle of 94°C for 3 min, followed by 35 cycles of 94°C for 30 s, 58°C for 30 s and 72°C for 2 min, and a final extension step of 72°C for 5 min. The conditions for the secondary PCR were the same except for 45 cycles instead of 35. All amplicons were visualised on a 1.5% agarose gel and bands were cut and purified using an in-house filter tip method defined by Yang et al. (2013).
For all potoroid species, amplicons were generated at the cox1 locus using the external primers COIF1 (5′-GGT TCA GGT GTT GGT TGG AC-3′) and COIF2 (5′-TAA GTA CAT CCC TAA TGT C-3′) and the internal primers COIbR1 (5′-CCA AGA GAT AAT ACR AAR TGG AA-3′) and COIbR2 (5′-ATA GTA TGT ATC ATG TAR WGC AA-3′) (Yang et al., 2016a). The PCR reactions and conditions were the same as per the 18S PCR carried out for woylie samples.
Sequencing and phylogenetic analysis
Purified amplicons were sequenced using an ABI PrismTM Dye Terminator Cycle Sequencing Kit (Applied Biosystems, California, USA) according to the manufacturer’s instructions. Samples were sequenced in the forward and reverse direction and the denaturation step was extended to 10 min to allow efficient primer binding. The sequences were analysed in Geneious v.8.1 (Kearse et al., 2012) and aligned using reference libraries with the MUSCLE (Edgar, 2004) plugin for Geneious v.8.1. All novel sequences were deposited in GenBank.
Using Bayesian methods, phylogenetic analyses were conducted for both gene regions (18S and cox1, respectively), to determine the evolutionary lineage for E. woyliei n. sp. (MK182524; MK202806), E. gaimardi (MK182525; MK202809), E. mundayi (MK182526; MK202808) and E. potoroi (MK182527; MK202807). JModelTest (Posada, 2008) was used to determine the most appropriate nucleotide substitution method for the Bayesian analyses for each gene region, which was the GTR+I+G (general time reversible gamma proportion of invariant sites) method. The Bayesian posterior probabilities were generated using MrBayes v.3.1.2 (10,000,000 generations, sampling frequency of 1,000, ‘burn in’ 3,000).
Results
The prevalence of coccidial infection in woylies is summarised by site in Table 1. It is important to note that Dryandra contains both resident woylies (endemic to the region), and translocated woylies originating from the Upper Warren region (specifically Balban, Boyicup, Corbal, Dudijup, Dwalgan and Winnejup). Likewise, Walcott and Warrup East contain both resident and translocated (originating from Perup Sanctuary) woylies. In the eastern bettong, 2 out of 4 (50%) samples were positive for E. gaimardi. In the long-nosed potoroo, 3 out of 12 (25.0%) samples were positive for E. potoroi, while 4 out of 12 (33.3%) samples were positive for E. mundayi; mixed infections with both coccidian parasites were identified in 2 out of 12 (16.7%) samples.
Family Eimeriidae Minchin, 1903
Genus Eimeria Schneider, 1875
Eimeria woyliei n. sp.
Type-host: Bettongia penicillata Gray (Mammalia: Marsupialia: Potoroidae), woylie or brush-tailed bettong.
Type-locality: Perup Sanctuary (34.16°S, 116.56°E) in the Upper Warren region, Western Australia, Australia.
Other localities: Corbal (34.10°S, 116.48°E), Dwalgan (34.07°S, 116.46°E) and Winnejup (34.07°S, 116.35°E) in the Upper Warren region, Western Australia, Australia; *Dryandra Woodland (32.80°S, 116.89°E) in the wheatbelt region, Western Australia, Australia (*translocated host, origin Dwalgan).
Type-material: Oöcysts in 10% formalin and oöcyst photosyntypes have been deposited in the Western Australian museum [reference numbers WAM Z91161 (holotype) and WAM Z91162-64 (paratypes)].
Prevalence: 21.2% (32 out of 151 specimens of the type-host; origin Perup Sanctuary).
Sporulation: Unknown.
Site of infection: Unknown, oöcysts recovered from faeces.
Representative DNA sequences: DNA sequences have been deposited in the GenBank sequence database under accession numbers MK182524-MK182527 (18S rRNA gene) and MK202806-MK202809 (cox1 gene).
Etymology: The name woyliei reflects the host species local name ‘woylie’, which is the Aboriginal name given to this animal by the Noongar people of south-western Western Australia (Abbott, 2001).
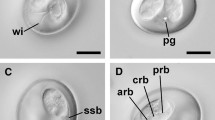
Description (Figs. 1,2A–C)
Sporulated oöcyst
Photomicrographs of sporulated oöcysts of Eimeria woyliei n. sp. (A–C) and a sporulated oöcyst of Eimeria gaimardi (D). Abbreviations: PG, polar granule; MP, micropyle; RB, refractile body; SB/SBB, Stieda body/sub-Stieda body; SMB, submicropyle body; SP, sporocyst; SR, sporocyst residuum. Scale-bars: 10 µm
Oöcysts (n = 86) pyriform-shaped [length 31.6–40.8 (36.7), width 22.6–31.0 (26.3), length/width (L/W) ratio 1.3–1.5 (1.4)] with smooth to slightly mammilate, bi-layered oöcyst wall, 1.3–1.7 (1.6) thick. Oöcyst wall thins at apex with visible micropyle. An irregular dome-like structure (referred to here as a submicropyle body) visible beneath micropyle. Polar granule irregular-shaped, apparent, although not detected in all sporulated oöcysts. Oöcyst residuum absent.
Sporocyst and sporozoite
Sporocysts (n = 76) ellipsoidal [length 13.3–17.8 (15.6), width 9.0–12.3 (10.3), L/W ratio 1.1–1.7 (1.5)]. Each sporocyst contains an indistinct dome-like Stieda body/sub-Stieda body. Sporocyst residuum present between 2 broadly elongate sporozoites. Sporozoites contain 2 distinct refractile bodies, one large (5–7 wide) and one small (up to 3 wide). Para-Stieda body absent.
Genetic characterisation of Eimeria spp. in potoroids
The phylogeny of the four potoroid Eimeria spp. was investigated using Bayesian analyses at two gene loci (18S and cox1). An alignment was generated for the 18S rDNA locus (1,158 bp, Fig. 3) and two alignments for the cox1 locus (686 bp, Fig. 4; 211 bp, Fig. 5). All alignments contained the novel sequences belonging to the four potoroid species as well as reference sequences downloaded from GenBank including an outgroup (Toxoplasma gondii). Similar phylogenetic relationships were observed for both loci, although some species were not observed in the cox1 tree due to lower availability of genetic data. A second cox1 alignment was generated (211 bp) in order to incorporate relevant marsupial species of smaller fragment size.
18S rDNA locus
Eimeria woyliei shared 99.2% genetic similarity with E. setonicis, isolated from the quokka Setonix brachyurus (Quoy & Gaimard) (KF225639; Austen et al., 2014), and 99.1% genetic similarity with E. trichosuri, from the mountain brushtail possum Trichosurus cunninghami Lindenmayer, Dubach & Viggers (FJ8292320; Power et al., 2009). Of the potoroid Eimeria spp., E. woyliei shared 98.8% genetic similarity with E. mundayi and E. potoroi, and 98.4% similarity with E. gaimardi.
cox1 locus
Using the longer (686 bp) cox1 locus, E. woyliei shared 97.1% similarity with E. trichosuri (JN192136; Ogedengbe et al., 2015), 95.7% similarity with E. potoroi, 95.3% similarity with E. gaimardi and 95.0% similarity with E. mundayi. Using the shorter (211 bp) cox1 locus, E. woyliei shared 94.8% similarity with Eimeria kanyana Bennett, Woolford, O’Hara, Nicholls, Warren & Hobbs, 2006, from the quenda Isoodon obesulus (Shaw) (KU845563/64; Hillman, 2016), and 97.2%, 96.7% and 95.3% similarity with E. potoroi, E. gaimardi and E. mundayi, respectively.
Discussion
Five species of Eimeria have been previously described in potoroid marsupials (Barker et al., 1988; Hulst et al., 2016). Of these, E. woyliei oöcysts most closely resemble E. gaimardi oöcysts from the eastern bettong in shape and size (range: 31.6–40.8 × 22.6–31.0 vs 32.0–39.2 × 20.8–26.4 µm; mean: 36.7 × 26.3 vs 34.6 × 24.3 µm) (see Barker et al., 1988). Sporocyst size of E. woyliei and E. gaimardi is also similar (range: 13.3–17.8 × 9.0–12.3 vs 13.6–16.0 × 9.0–10.4 µm; mean: 15.6 × 10.3 vs 15.0 × 9.6 µm). However, oöcysts and sporocysts in E. woyliei are slightly larger on average. Eimeria woyliei oöcysts also contain a micropyle, submicropyle body and a polar granule; E. gaimardi oöcysts do not according to the original morphological description by Barker et al. (1988). Based on the sporulated E. gaimardi oöcysts examined during this study however, we propose that E. gaimardi oöcysts do possess a micropyle, submicropyle body and polar granule (Fig. 2D). Thus, the major distinguishing morphological characteristic between E. woyliei and E. gaimardi is the appearance of the oöcyst wall, which for E. gaimardi appears mammilated and more robust (Fig. 2D). The oöcyst wall of E. woyliei in comparison is smooth to slightly mammilate.
Eimeria woyliei oöcysts are noticeably larger than E. potoroi oöcysts [24.0–29.6 × 16.8–22.4 (26.2 × 18.5) µm] (Barker et al., 1988) and E. mundayi oöcysts [13.6–20.8 × 13.6–19.2 (16.9 × 16.2) µm] from the long-nosed potoroo (Barker et al., 1988). Eimeria woyliei oöcysts are comparable in size to E. aepyprymni oöcysts [32.0–42.8 × 18.4–25.2 (36.7 × 21.9) µm] from the rufous bettong Aepyprymnus rufescens (Gray) (Barker et al., 1988); however, E. aepyprymni oöcysts are ovoid rather than pyriform. While similar in shape (pyriform), E. woyliei oöcysts are considerably larger than E. burdi oöcysts [21.0–24.0 × 14.0–16.0 (22.6 × 14.9) µm] from the burrowing bettong Bettongia lesueur (Quoy & Gaimard) (Hulst et al., 2016).
Using the 18S rDNA locus, E. woyliei was grouped within the marsupial clade and was most similar to E. setonicis from the quokka. Eimeria woyliei oöcysts can be morphologically distinguished from oöcysts of E. setonicis by shape (pyriform vs ellipsoidal) (Austen et al., 2014). Despite the morphological similarity between oöcysts of E. woyliei and E. gaimardi, E. woyliei was more genetically similar to E. mundayi and E. potoroi, rather than E. gaimardi, though this difference was minimal.
Using the cox1 locus, E. woyliei was most similar to E. trichosuri from the mountain brushtail possum and grouped within the same clade as E. macropodis from the tammar wallaby Macropus eugenii (Desmarest) (Hill et al., 2012), and E. kanyana from the quenda (Hillman et al., 2016). Eimeria woyliei oöcysts can be differentiated from oöcysts of E. trichosuri by shape (pyriform vs ellipsoidal; Power et al., 2009); E. trichosuri oöcysts are also larger [34.4–49.2 × 18.4–27.8 (41.4 × 22.7) µm] (O’Callaghan & O’Donoghue, 2001). Unexpectedly, Isospora amphiboluri Canon, 1967, from the central bearded dragon Pogona vitticeps (Ahl) was also present within the same phylogenetic clade (GenBank: KR108297). Although cox1 sequences are deemed superior to nuclear 18S rDNA sequences for delimiting species (Ogedengbe et al., 2011), cox1 sequences on GenBank are limited. Nonetheless the supposed relationship between E. woyliei and I. amphiboluri remains unclear.
Within Australia various coccidial species (Eimeria and less commonly Isospora) have been recorded in captive and free-ranging macropods (Mykytowycz, 1964; Barker et al., 1989; Yang et al., 2012; Duszynski, 2016). Traditional morphological criteria have been useful for identifying coccidial species infecting wildlife. However, the implementation of genetic characterisation combining both 18S rDNA and cox1 loci helps to discriminate between morphologically similar species and provides an accurate measure of the evolutionary relationship between coccidial species (Power et al., 2009; Yang et al., 2012). As some Eimeria spp. are capable of infecting more than one marsupial host (Barker et al., 1989) and marsupials tend to harbour multiple Eimeria spp. (Power et al., 2009), this knowledge may be useful for predicting potential avenues of disease spread during the management of threatened populations (e.g. during fauna translocation). This study contributes toward our knowledge of Eimeria spp. infecting potoroid marsupials. Eimeria woyliei parasitising woylies is the sixth Eimeria spp. to be formally described from potoroid marsupials and we have genetically characterised four of the six known potoroid Eimeria species.
References
Abbott, I. (2001). Aboriginal names of mammal species in south-west Western Australia. CALMscience, 3, 433–486.
Austen, J. M., Friend, J. A., Yang, R., & Ryan, U. M. (2014). Further characterisation of two Eimeria species (Eimeria quokka and Eimeria setonicis) in quokkas (Setonix brachyurus). Experimental Parasitology, 138, 48–54.
Barker, I. K., Harrigan, K. E., & Dempster, J. K. (1972). Coccidiosis in wild grey kangaroos. International Journal for Parasitology, 2, 187–192.
Barker, I. K., O’Callaghan, M. G., & Beveridge, I. (1988). Eimeria spp. (Apicomplexa: Eimeriidae) parasitic in the rat-kangaroos Hypsiprymnodon moschatus, Potorous tridactylus, Aepyprymnus rufescens and Bettongia gaimardi (Marsupialia: Potoroidae). International Journal for Parasitology, 18, 947–953.
Barker, I. K., O’Callaghan, M. G., & Beveridge, I. (1989). Host-parasite associations of Eimeria spp. (Apicomplexa: Eimeriidae) in kangaroos and wallabies of the genus Macropus (Marsupialia: Macropodidae). International Journal for Parasitology, 19, 241–263.
Duszynski, D. W. (2016). The Biology and Identification of the Coccidia (Apicomplexa) of Marsupials of the World (1st edn.). London, UK: Elsevier Inc., pp. 31–86.
Edgar, R. C. (2004). MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Research, 32, 1792–1797.
Hill, N. J., Richter, C., & Power, M. L. (2012). Pinning down a polymorphic parasite: new genetic and morphological descriptions of Eimeria macropodis from the Tammar wallaby (Macropus eugenii). Parasitology International, 61, 461–465.
Hillman, A. E., Yang, R., Lymbery, A. J., & Thompson, R. C. A. (2016). Eimeria spp. infecting quenda (Isoodon obesulus) in the greater Perth region. Western Australia. Experimental Parasitology, 170, 148–155.
Hing, S., Northover, A. S., Narayan, E. J., Wayne, A. F., Jones, K. L., Keatley, S., et al. (2017). Evaluating stress physiology and parasite infection parameters in the translocation of critically endangered woylies (Bettongia penicillata). EcoHealth, 14, 1–11.
Hulst, F., Kemp, L. F., & Slapeta, J. (2016). A new coccidian parasite of the boodie, Bettongia lesueur (Mammalia: Marsupialia: Potoroidae), from Australia. Folia Parasitologica, 63, 1–4.
Kearse, M., Moir, R., Wilson, A., Stones-Havas, S., Cheung, M., Sturrock, S., et al. (2012). Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics, 28, 1647–1649.
Mykytowycz, R. (1964). Coccidia in wild populations of the red kangaroo, Megaleia rufa (Desmarest), and the grey kangaroo, Macropus canguru (Muller). Parasitology, 54, 105–115.
Northover, A. S., Godfrey, S. S., Lymbery, A. J., Morris, K., Wayne, A. F., & Thompson, R. C. A. (2017). Evaluating the effects of ivermectin treatment on communities of gastrointestinal parasites in translocated woylies (Bettongia penicillata). EcoHealth, 14, 117–127.
O’Callaghan, M. G., & O’Donoghue, P. J. (2001). A new species of Eimeria (Apicomplexa: Eimeriidae) from the brushtail possum, Trichosuris vulpecula (Diprotodontia: Phalangeridae). Transactions of the Royal Society of South Australia, 125, 129–132.
Ogedengbe, J. D., Hanner, R. H., & Barta, J. R. (2011). DNA barcoding identifies Eimeria species and contributes to the phylogenetics of coccidian parasites (Eimeriorina, Apicomplexa, Alveolata). International Journal for Parasitology, 41, 843–850.
Ogedengbe, J. D., Ogedengbe, M. E., Hafeez, M. A., & Barta, J. R. (2015). Molecular phylogenetics of eimeriid coccidia (Eimeriidae, Eimeriorina, Apicomplexa, Alveolata): A preliminary multi-gene and multi-genome approach. Parasitology Research, 114, 4149–4160.
Power, M. L., Richter, C., Emery, S., Hufschmid, J., & Gillings, M. R. (2009). Eimeria trichosuri: Phylogenetic position of a marsupial coccidium, based on 18S rDNA sequences. Experimental Parasitology, 122, 165–168.
Posada, D. (2008). jModelTest: Phylogenetic model averaging. Molecular Biology and Evolution, 25, 1253–1256.
Vogelnest, L., & Portas, T. (2008). Chapter 7: Macropods.In: Vogelnest, L. & Woods, R. (Eds), Medicine of Australian Mammals (1st edn). Collingwood: CSIRO Publishing, pp. 196–197.
Yang, R., Fenwick, S., Potter, A., Elliot, A., Power, M., Beveridge, I., et al. (2012). Molecular characterization of Eimeria species in macropods. Experimental Parasitology, 132, 216–221.
Yang, R., Murphy, C., Song, Y., Ng-Hublin, J., Estcourt, A., Hijjawi, N., et al. (2013). Specific and quantitative detection and identification of Cryptosporidium hominis and C. parvum in clinical and environmental samples. Experimental Parasitology, 135, 142–147.
Yang, R., Brice, B., & Ryan, U. (2016a). Morphological and molecular characterization of Choleoeimeria pogonae n. sp. coccidian parasite (Apicomplexa: Eimeriidae, 1989, Paperna and Landsberg) in a western bearded dragon (Pogona minor minor). Experimental Parasitology, 160, 11–16.
Yang, R., Brice, B., & Ryan, U. (2016b). Morphological and molecular characterization of Eimeria purpureicephali n. sp. (Apicomplexa: Eimeriidae) in a red-capped parrot (Purpureicephalus spurius Kuhl, 1820) in Western Australia. International Journal for Parasitology: Parasites and Wildlife, 5, 34–39.
Acknowledgements
We would like to thank Julia Wayne, Marika Maxwell, Colin Ward, Chris Vellios, Peter Wnuk, Malcolm Ovans, Brian Macmahon and other DBCA staff for their assistance in the field, and all of our dedicated volunteers for their support with fieldwork, sample collection and processing. We would also like to express our gratitude to the staff from Bonorong Wildlife Sanctuary; Elise Dewar, Micah Visoiu and Michael Driessen from DPIPWE; and John Lawson from Dryandra Woodland, for their assistance with faecal sample collection.
Funding
This study was principally funded by the Australian Research Council (LP130101073). We have also been supported by grants from the Holsworth Wildlife Research Endowment (HOLSW2015-1-F149) and the Ecological Society of Australia, The Royal Zoological Society of New South Wales (Paddy Pallin Grant) and the Australian Wildlife Society.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
All applicable international, national, and/or institutional guidelines for the care and use of animals were followed. Samples from the woylie were collected under DBCA Scientific Licenses (Regulation 4: written notice of lawful authority; and 17: licence to take fauna for scientific purposes) and with approval from the Murdoch University Animal Ethics Committee (RW2659/14). Samples from the eastern bettong were collected with permission from Bonorong Wildlife Sanctuary. Samples from the long-nosed potoroo were collected under authorities and permits issued to the Department of Primary Industries, Parks, Water and Environment (DPIPWE) staff to live-trap wildlife on reserved land in Tasmania, following the Standard Operating Procedures for Live-trapping and Handling of Wild Tasmanian Mammals 2013 by the DPIPWE.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
This article was registered in the Official Register of Zoological Nomenclature (ZooBank) as 01B2910A-6B10-40B0-ABB3-40725DDA9FD4. This article was published as an Online First article on the online publication date shown on this page. The article should be cited by using the doi number. This is the Version of Record.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Northover, A.S., Keatley, S., Elliot, A.D. et al. Identification of a novel species of Eimeria Schneider, 1875 from the woylie, Bettongia penicillata Gray (Diprotodontia: Potoroidae) and the genetic characterisation of three Eimeria spp. from other potoroid marsupials. Syst Parasitol 96, 553–563 (2019). https://doi.org/10.1007/s11230-019-09870-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11230-019-09870-y