Abstract
In unsupervised classification, Hidden Markov Models (HMM) are used to account for a neighborhood structure between observations. The emission distributions are often supposed to belong to some parametric family. In this paper, a semiparametric model where the emission distributions are a mixture of parametric distributions is proposed to get a higher flexibility. We show that the standard EM algorithm can be adapted to infer the model parameters. For the initialization step, starting from a large number of components, a hierarchical method to combine them into the hidden states is proposed. Three likelihood-based criteria to select the components to be combined are discussed. To estimate the number of hidden states, BIC-like criteria are derived. A simulation study is carried out both to determine the best combination between the combining criteria and the model selection criteria and to evaluate the accuracy of classification. The proposed method is also illustrated using a biological dataset from the model plant Arabidopsis thaliana. A R package HMMmix is freely available on the CRAN.







Similar content being viewed by others
References
Baudry, J.P., Raftery, A.E., Celeux, G., Lo, K., Gottardo, R.: Combining mixture components for clustering. J. Comput. Graph. Stat. 9(2), 332–353 (2010). doi:10.1198/jcgs.2010.08111
Bérard, C., Martin-Magniette, M.-L., Robin, S.: Mixture model approach to compare two samples of tiling array data: chip-chip and transcriptome. Stat. Appl. Genet. Mol. Biol. 10(1), 1–22 (2011). doi:10.2202/1544-6115.1692
Biernacki, C., Celeux, G., Govaert, G.: Assessing a mixture model for clustering with the integrated completed likelihood. IEEE Trans. Pattern Anal. Mach. Intell. 22, 719–725 (2000). ISSN 0162-8828
Cappé, O., Moulines, E., Ryden, T.: Inference in Hidden Markov Models. Springer, Berlin (2010)
Celeux, G., Durand, J.B.: Selecting hidden Markov model state number with cross-validated likelihood. In: Computational Statistics, pp. 541–564 (2008)
Chatzis, S.P.: Hidden Markov models with nonelliptically contoured state densities. IEEE Trans. Pattern Anal. Mach. Intell. 32, 2297–2304 (2010). ISSN 0162-8828
Dempster, A.P., Laird, N.M., Rubin, D.B.: Maximum likelihood from incomplete data via the em algorithm. J. R. Stat. Soc. B 39(1), 1–38 (1977)
Durbin, R., Eddy, S.R., Krogh, A., Mitchison, G.: Biological Sequence Analysis: Probabilistic Models of Proteins and Nucleic Acids. Cambridge University Press, Cambridge (1998)
Fraley, C., Raftery, A.E.: Mclust: software for model-based cluster analysis. J. Classif. 16, 297–306 (1999)
Hennig, C.: Methods for merging Gaussian mixture components. Adv. Data Anal. Classif. 4, 3–34 (2010). doi:10.1007/s11634-010-0058-3
Keribin, C.: Consistent estimation of the order of mixture models. Sankhya, Ser. A 62, 49–66 (2000). doi:10.2307/25051289
Li, J.: Clustering based on a multilayer mixture model. J. Comput. Graph. Stat. 14(3), 547–568 (2005). doi:10.1198/106186005X59586
Li, W., Meyer, A., Liu, X.S.: A hidden Markov model for analyzing chip-chip experiments on genome tiling arrays and its application to p53 binding sequences. Bioinformatics 211, 274–282 (2005)
Lippman, Z., Gendreland, A.-V., Blackand, M., Vaughn, M.W., et al.: Role of transposable elements in heterochromatin and epigenetic control. Nature 430, 471–476 (2004)
McLachlan, G.J., Peel, D.: In: Finite Mixture Models (2000)
Rabiner, L.R.: A tutorial on hidden Markov models and selected applications in speech recognition. Proc. IEEE 77(2), 257–286 (1989). doi:10.1109/5.18626
Roudier, F., Ahmed, I., Bérard, C., Sarazin, A., Mary-Huard, T., et al.: Integrative epigenomic mapping defines four main chromatin states in arabidopsis. EMBO J. 30, 1928–1938 (2011)
Schwarz, G.: Estimating the dimension of a model. Ann. Stat. 6, 461–464 (1978)
Sun, W., Cai, T.: Large-scale multiple testing under dependence. J. R. Stat. Soc. 71, 393–424 (2009)
Author information
Authors and Affiliations
Corresponding author
Appendix
Appendix
1.1 A.1 Algorithm
We present in this appendix the algorithm we proposed for combining components of an HMM. This algorithm has been written with respect to the results we obtained in Sect. 4.1.2. However, this algorithm can easily be written for other criteria.
-
1.
Fit an HMM with K components.
-
2.
From G=K,K−1,…,1
-
Select the clusters k and l to be combined as:

-
Update the parameters with a few steps of the EM algorithm to get closer to a local optimum.
-
-
3.
Selection of the number of groups \(\widehat{D}\):

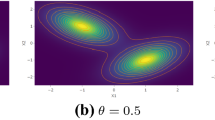
1.2 A.2 Mean and variance of the Gaussian distributions for the simulation study (Sect. 4.1)

and,

Rights and permissions
About this article
Cite this article
Volant, S., Bérard, C., Martin-Magniette, ML. et al. Hidden Markov Models with mixtures as emission distributions. Stat Comput 24, 493–504 (2014). https://doi.org/10.1007/s11222-013-9383-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11222-013-9383-7