Abstract
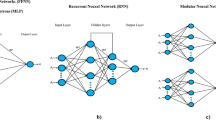
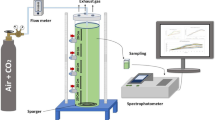
This work was aimed at applying machine learning techniques to predict Semi-Solid Fermentation (SSF) yield using Cunninghamella echinulata fungus for alginate lyase production using Sargassum macroalgae genus as substrate. Approximately 115 experimental data were obtained as a result of the variation of the following factors: tc (cultivation time) (1–7 days), Ci (inoculum concentration) (2.106–1.107 spore/gbiomass), pHNutri (nutrient solution pH) (2.5–8.5), uw/w (moisture content) (65–85% w/w) and %inducer (sodium alginate concentration) (0–33.33% w/w), which were the input variables and alginate lyase enzyme concentration (U mL−1) as the output variable. The programming language employed was Python, through its SciPylibrary. The non-linear relationship between fermentation, the factors and objectives were determined using the learning capacity of Artificial Neural Networks (ANN) and the Support Vector Machine (SVM), through the correlation coefficient (R2). For ANN, with 15 neurons and using the logistic activation function, a R2 = 0.877 was obtained. SVM, with a polynomial kernel function obtained R2 = 0.821. In both models, the variables with influence were tc, uw/w, %inducer and pHNutri. Ci did not have any significant influence in the range studied. In both methods, the R2 found was considered satisfactory contributing to the optimization of the alginate lyase production using Cunninghamella echinulata.


Similar content being viewed by others
References
Guiry MD (2014) AlgaeBase. World-wide eletronic publication. National University of Ireland, Galway. http://www.algaebase.org. Accessed 8 Sept 2021
Torres MR, Sousa APA, Silva Filho EAT et al (2007) Extraction and physicochemical characterization of Sargassum vulgare alginate from Brazil. Carbohydr Res 342(14):2067–2074. https://doi.org/10.1016/j.carres.2007.05.022
Karina T (2017) Estudo decifra mistério das algas que invadiram o Brasil. https://exame.com/ciencia/estudo-decifra-misterio-das-algas-que-invadiram-o-brasil/#:~:text=Embora%20as%20algas%20do%20g%C3%AAnero,o%20banho%20de%20mar%20invi%C3%A1vel. Accessed 07 Sept 2021
Trinanes J, Putman NF, Goni G et al (2021) Monitoring pelagic Sargassum inundation potential for coastal communities. J Oper Oceanogr 1:1–12. https://doi.org/10.1080/1755876X.2021.1902682
Bonilla-Loaiza AM, Rodríguez-Jasso RM, Belmares R et al (2022) Fungal Proteins from Sargassum spp. using solid-state fermentation as a green bioprocess strategy. Molecules 27(12):3887. https://doi.org/10.3390/molecules27123887
Chavez-Gonzalez ML, Balagurusamy N, Aguilar CN (2019) Advances in Food Bioproducts and Bioprocessing Technologies. CRC Press, Boca Raton
Mohapatra BR (2020) Biocatalytic characteristics of chitosan nanoparticle-immobilized alginate lyase extracted from a novel Arthrobacter species AD-10. Biocatal Agric Biotechnol 23:101458. https://doi.org/10.1016/j.bcab.2019.101458
Siller-Sánchez A, Ruiz HA, Aguilar CN et al (2019) Green bio-processes. In: Parameswaran B, Varjani S, Raveendran S (eds) Biorefinery approach for red seaweeds biomass as source for enzymes production: food and biofuels industry. Springer, Singapore, pp 413–446
Lara A, Rodríguez-Jasso RM, Loredo-Treviño A et al (2020) Enzymes in the third generation biorefinery for macroalgae biomass. Elsevier, Amsterdam
Sivakumar T, Sathya C, Shankar T et al (2015) Screening and optimization of alginate lyase producing Bacillus sp. from seaweed. ANJAC J Sci 14:42–429
Beltagy EA, El Borai A, Lewiz M et al (2016) Purifcation and characterization of alginate lyase from locally isolated marine Pseudomonas stutzeri MSEA04. Acta Biol Hung 67(3):305–317. https://doi.org/10.1556/018.67.2016.3.8
Li S, Wang L, Hao J et al (2017) Purification and characterization of a new alginate lyase from marine bacterium Vibrio sp. SY08. Mar Drugs 15(1):1. https://doi.org/10.3390/md15010001
Shankar T, Sivakumar T, Satya C et al (2016) Purification, characterization and immobilization of alginate lyase produced by Bacillus sp. associated with Sargassum wightii. Univers J Microbiol Res 4(1):11–22. https://doi.org/10.13189/ujmr.2016.040103
Zhu Y, Wu L, Chen Y et al (2016) Characterization of na extracelular biofunctional alginate lyase from marine Microbulbifer sp. ALW1 and antioxidante activity of enzymatic hydrolysates. Microbiol Res 182:49–58. https://doi.org/10.1016/j.micres.2015.09.004
Tavafi H, Abdi-Ali AA, Ghadam P et al (2017) Screening and optimization of media compositions for extracelular alginate lyase production. Iran Biomed J 21(1):48–56. https://doi.org/10.6091/.21.1.48
Wang X, Chen J, Liu C et al (2010) Hybrid modeling of penicillin fermentation process based on least square support vector machine. Chem Eng Res Des 88(4):415–420. https://doi.org/10.1016/j.cherd.2009.08.010
Chen P, Zhu Y, Men Y et al (2018) Purification and characterization of a novel alginate lyase from the marine bacterium Bacillus sp. Alg07. Mar Drugs 16(3):86. https://doi.org/10.3390/md16030086
Hifney AF, Fawzy MA, Abdel-Gawad KM et al (2018) Upgrading the antioxidante properties of fucoidan and alginate from Cystoseira trinodis by funfgal fermentation or enzymatic pretreatment of the seaweed biomass. Food Chem 269:387–395. https://doi.org/10.1016/j.foodchem.2018.07.026
Li S, Wang ZP, Wang LN et al (2019) Combined enzymatic hydrolysis and selective fermentation for green production of alginate oligosaccharides from Laminaria japonica. Bioresour Technol 281:84–89. https://doi.org/10.1016/j.biortech.2019.02.056
Sun X, Shen W, Gao Y et al (2019) Heterologous expression and purification of a marine alginate lyase in Escherichia coli. Protein Expr Purif 153:97–104. https://doi.org/10.1016/j.pep.2018.09.002
Qiao L, Yang X, Xie R et al (2020) Efficient production of ulvan lyase from Ulva prolifera by Catenovulum sp. LP based on stage-controlled fermentation strategy. Algal Res 46:101812. https://doi.org/10.1016/j.algal.2020.101812
Ruiz HA, Rodríguez-Jasso RM, Rodríguez R et al (2012) Pectinase production from lemon peel pomace as support and carbon source in solid-state fermentation column-tray bioreactor. Biochem Eng J 65:90–95. https://doi.org/10.1016/j.bej.2012.03.007
Gervais P, Molin P (2003) The role of water in solid state fermentation. Biochem Eng J 13(2–3):85–101. https://doi.org/10.1016/S1369-703X(02)00122-5
Sun Y, Cheng J (2002) Hydrolysis of lignocellulosic materials for ethanol production: a review. Bioresour Technol 83(1):1–11. https://doi.org/10.1016/S0960-8524(01)00212-7
Coelho MAZ, Salgado AM, Ribeiro BD (2018) In: EPUB (ed) Tecnologia Enzimática, 10 edn. FAPERJ, Petrópolis
Merino A, Eibes G, Hormaza A (2019) Effect of copper and different carbon and nitrogen sources and the decolorization of an industrial dye mixture under solid-state fermentation. J Clean Prod 237:117713. https://doi.org/10.1016/j.jclepro.2019.117713
Silva FL, Campos AO, Santos DA et al (2018) Pretreatments of Carnauba (Copernicia prunifera) straw residue for production of cellulolytic enzymes by Trichoderma reesei CCT-2768 by solid state fermentation. Renew Energy 116(Part A):299–308. https://doi.org/10.1016/j.renene.2017.09.064
Aita BC, Spannemberg SS, Schmaltz S et al (2019) Production of cell-wall degrading enzymes by solid-state fermentation using agroindustrial residues as substrates. J Enviro Chem Eng 7(3):103193. https://doi.org/10.1016/j.jece.2019.103193
Bailey JE (1998) Mathematical modeling and analysis in biochemical engineering: past accomplishments and future opportunities. Biotechnol Prog 14(1):8–20. https://doi.org/10.1021/bp9701269
Torres NV, Santos G (2015) The (Mathemayical) modeling process in biosciences. Front Genet 6:354. https://doi.org/10.3389/fgene.2015.00354
Van Can HJL, Hellinga C, Luyben KCA et al (1996) Strategy for dynamic process modelling based on neural networks in macroscopic balances. AIChE J 42(12):3403–3418. https://doi.org/10.1002/aic.690421211
Adorno WT, Martins GAS, Silva WG (2013) Modelagem matemática aplicada á transferência de massa em alimento. Enciclopédia Biosfera 9:1465–1478
Lek S, Guegan JF (1999) Artificial neural networks as a tool in ecological modelling, an introduction. Ecol Model 120(2–3):65–73. https://doi.org/10.1016/S0304-3800(99)00092-7
Larrañaga P, Calvo B, Santana R et al (2006) Machine learning in bioinformatics. Brief Bioinform 7(1):86–112. https://doi.org/10.1093/bib/bbk007
Fabris F, Magalhães JPD, Freitas AA (2017) A review of supervised machine learning applied to ageing research. Biogerontology 18(2):171–188. https://doi.org/10.1007/s10522-017-9683-y
Kotsiantis SB, Zaharakis ID, Pintelas PE (2006) Machine learning: a review of classification and combining techniques. Artif Intell Ver 26:159–190. https://doi.org/10.1007/s10462-007-9052-3
Vieira WF, Correa HT, Campos ES et al (2020) A novel multiple reactor system for the long-term production of L-asparaginase by Penicillium sp. LAMAI 505. Process Biochem 90:23–31. https://doi.org/10.1016/j.procbio.2019.11.012
Heckmann D, Schlüter U, Weber APM (2017) Machine learning techniques for predictingcrop photosynthetic capacity from leaf reflectance spectra. Mol Plant 10(6):878–890. https://doi.org/10.1016/j.molp.2017.04.009
Azuaje F (2016) Computational models for predicting drug responses in cancer research. Brief Bioinform 18(5):820–829. https://doi.org/10.1093/bib/bbw065
Barchi AC, Ito S, Escaramboni B et al (2016) Artificial intelligence approach based on near-infrared spectral data for monitoring of solid-state fermentation. Process Biochem 51(10):1338–1347. https://doi.org/10.1016/j.procbio.2016.07.017
Malhis N, Wong ET, Nassar R et al (2015) Computational identification of MoRFs in protein sequences using hierarchical application of Bayes Rule. PLoS ONE 10(10):e0141603. https://doi.org/10.1371/jornal.pone.0141603
Kandoi G, Acencio ML, Lemke N (2015) Prediction of druggable proteins using machine learning and systems biology: a mini-review. Front Physiol 6:366. https://doi.org/10.3389/fphys.2015.00366
Pappu SMJ, Gummadi SN (2017) Artificial neural network and regression coupled genetic algorithm to optimize parameters for enhanced xylitol production by Debaryomyces nepalensis in bioreactor. Biochem Eng J 120:136–145. https://doi.org/10.1016/j.bej.2017.01.010
Hanai T, Honda H, Ohkusu E et al (1999) Application of an artificial neural network and genetic algorithm for determination of process orbits in the koji making process. J Biosci Bioeng 87(4):507–512. https://doi.org/10.1016/s1389-1723(99)80101-7
Craninx M, Fievez V, Vlaeminck B et al (2008) Artificial neural network models of the rumen fermentation pattern in dairy cattle. Comput Electron Agr 60(2):226–238. https://doi.org/10.1016/j.compag.2007.08.005
Desai KM, Vaidya BK, Singhal RS et al (2005) Use of an artificial neural network in modeling yeast biomass and yield of β-glucan. Process Biochem 40(5):1617–1626. https://doi.org/10.1016/j.procbio.2004.06.015
Zhu B, Ye S, Jiang M et al (2009) Achieving the carbon intensity target of china: a least squares support vector machine with mixture kernel function approach. Appl Energy 233–234:196–207. https://doi.org/10.1016/j.apenergy.2018.10.048
Zheng ZY, Guo XN, Zhu KX et al (2017) Artificial neural network—Genetic algorithm to optimize wheat germ fermentation condition: application to the production of two anti-tumor benzoquinones. Food Chem 227:264–270. https://doi.org/10.1016/j.foodchem.2017.01.077
Bao J, Zhang X, Zheng JH et al (2018) Mixed Fermentation of Spirulina platensis with Lactobacillus plantarum and Bacillus subtilis by Random-Centroid Optimization. Food Chem 264:64–72. https://doi.org/10.1016/j.foodchem.2018.05.027
Rayovarapu B, Tallopragada P, Usha MS (2019) Statistical optimization of γ-aminobutyric acid production by response surface methodology and artificial neural network models using Lactobacillus fermentum isolated from palm wine. Biocatal Agric Biotechnol 22:101362. https://doi.org/10.1016/j.bcab.2019.101362
Fischer J, Lopes VS, Cardoso SL et al (2017) Machine learning techniques applied to lignocellulosic ethanol in simultaneous hydrolysis and fermentation. Braz J Chem Eng 34(1):53–63. https://doi.org/10.1590/0104-6632.20170341s20150475
Kimutai G, Ngenzi A, Said RN et al (2020) An optimum tea fermentation detection model based on deep convolutional neural networks. Data 5(2):44. https://doi.org/10.3390/data5020044
Angelia RE, Linsangan N (2018) Classificação do nível de fermentação de grãos de cacau de corte cruzado usando o algoritmo k-NN. https: //doi.org/https://doi.org/10.1145/3309129.3309142. Accessed 8 Sept 2021
Zhu X, Zhu Z (2018) The generalized predictive control of bacteria concentration in marine lysozyme fermentation process. Food Sci Nutr 6(8):2459–2465. https://doi.org/10.1002/fsn3.850
Boggione JM, Allasia MB, Bassani G, Farruggia B (2016) Potential use of soybean hulls and waste paper as supports in SSF for celulase production by Aspergillus niger. Bioprocess Engi 6:1–8. https://doi.org/10.1016/j.bcab.2016.02.003
Abud AKS, Araújo ML, Almeida RMRG (2015) Uso de resíduo de laranja lima e de casca de coco verde na produção de enzimas. Scientia Plena 11(10):1–8. https://doi.org/10.14808/sci.plena.2015.104201
Growtthaman MK, Krishna C, Moo-Young M (2001) Fungal solid state fermentation —na overview. Agric Food Product 1:305–353. https://doi.org/10.1016/S1874-5334(01)80014-9
Miller JG (1959) Use of dinitrosalicyclic acid reagent for determination of reducing sugars. Anal Chem 31(3):426–428. https://doi.org/10.1021/ac60147a030
Swift SM, Hudgens JW, Heselpoth RD et al (2014) Characterization of AlgMsp, an alginate lyase from Microbulbifer sp. 6532A. PLoS ONE 9(11):e112939. https://doi.org/10.1371/jornal.pone.0112939
Saraiva SV, Amorim RFC, Carvalho FO et al (2014) Prognóstico de radiação solar através modelos que combinam as técnicas wavelet e redes neurais. Rev Bras Geogr Fís 7(5):808–817
Karri RR, Tanzifi M, Yaraki MT, Sahu JN (2018) Optimization and modeling of methyl orange adsorption onto polyaniline nano-adsorbent through response surface methodology and differential evolution embedded neural network. J Environ Manage 223:517–529. https://doi.org/10.1016/j.jenvman.2018.06.027
Géron A (2019) Hands-on machine learning with Scikit-Learn, Keras, and TensorFlow: concepts, tools, and techniques to build intelligent systems. O’Reilly Media Inc, Sebastopol
Reis CH (2018) Otimização de Hiperparâmetros em Redes Neurais Profundas. https://carlos-henreis.github.io/files/Monografia_TFG.pdf. Accessed 08 Sept 2021
Goodswen SJ, Barratt JL, Kennedy PJ et al (2021) Machine learning and applications in microbiology. FEMS Microbiol Rev 45(5):fuab015. https://doi.org/10.1093/femsre/fuab015
Saputro DRS, Widyaningsih P (2017) Limited memory Broyden-Fletcher-Goldfarb-Shanno (L-BFGS) method for the parameter estimation on geographically weighted ordinal logistic regression model (GWOLR). AIP Conf Proc 1868:040009. https://doi.org/10.1063/1.4995124
Murtagh F (1991) Multilayer perceptrons for classification and regression. Neurocomputing 2(5–6):183–197. https://doi.org/10.1016/0925-2312(91)90023-5
Silva RG, Cruz AJG, Hokka CO et al (2000) A hybrid feedforward neural network model for the cephalosporin C production process. Braz J Chem Eng 17:4–7. https://doi.org/10.1590/S0104-66322000000400023
Bouaoudat BD, Yalaoui F, Amodeo L et al (2012) Efficient developments in modeling and optimization of solid state fermentation. Biotechnol Biotechnol Equip 29(6):1216–1225. https://doi.org/10.5504/bbeq.2012.0108
Michel V, Gramfort A, Varoquaux G et al (2012) A supervised clustering approach for fMRI-based inference of brain states. Pattern Recognit 45(6):2041–2049. https://doi.org/10.1016/j.patcog.2011.04.006
Oliveira AVM, Silva MM, Castro AF (2020) Estudo Comparativo de Desempenho Entre SVM e MLP no Reconhecimento de Imagens. I Meeting of Computation of the Oeste Potiguar—Pocket. https://periodicos.ufersa.edu.br/ecop/article/view/10115. Accessed 07 Sept 2021
Palácios RHC, Silva IN, Goedtel A et al. (2013) Estimador de torque em motores de indução trifásicos com alimentação desequilibrada baseado em redes neurais artificiais. XX Brazilian Congress of Automatics. http://www.swge.inf.br/CBA2014/anais/PDF/1569934411.pdf Accessed 23 Aug 2021
Achirul NM, Boro Seminar K, Nandika D et al (2018) A Comparison Study of kernel functions in the support vector machine and its application for termite detection. Information 9(1):5. https://doi.org/10.3390/info9010005
Kasnavi SA, Aminafshar M, Shariati MM et al (2018) The effect of kernel selection on genome wide prediction of discrete traits by support vector machine. Gene Rep 11:279–282. https://doi.org/10.1016/j.genrep.2018.04.006
Hassan MAM, Xu S, Kabir MMJ et al (2016) Performance evaluation of different kernels for support vector machine used in intrusion detection system. J Comput Netw Commun 8(6):39–53. https://doi.org/10.5121/ijcnc.2016.8604
Chui KT, Lytras MD (2019) A novel MOGA-SVM multinomial classification for organ inflammation detection. Appl Sci 9(11):2284. https://doi.org/10.3390/app9112284
Saeed S, Ong HC (2019) Performance of SVM with Multiple Kernel Learning for Classification Tasks of Imbalanced Datasets. http://www.pertanika.upm.edu.my/resources/files/Pertanika%20PAPERS/JST%20Vol.%2027%20(1)%20Jan.%202019/30%20JST-1133-2018.pdf. Accessed 01 Sept 2021
Cervantes J, Garcia-Lamont F, Rodríguez-Mazahua L et al (2020) A comprehensive survey on support vector machine classification: applications, challenges and trends. Neurocomputing 408:189–215. https://doi.org/10.1016/j.neucom.2019.10.118
Shaikhina T, Lowe D, Daga S et al (2015) Machine learning for predictive modelling based on small data in biomedical engineering. IFAC-PapersOnLine 48(20):469–474. https://doi.org/10.1016/j.genrep.2018.04.006
Silva MCS, Silva CEF, Santos LM, Medeiros JA, Vieira RC, Abud AKS, Almeida RMRG, Tonholo J (2022) Alginate lyase produced by filamentous fungus through solid state fermentation using Sargassum from the Brazilian coast. Waste Biomass Valoriz. https://doi.org/10.1007/s12649-022-01706-z
Acknowledgements
The authors are grateful for the financial support by CNPq—Brazil (National Council of Research and Development of Brazil)—Process numbers 313195/2019-6, 440070/2019-8 and 407274/2018-9 and by calling 16/2020.
Author information
Authors and Affiliations
Contributions
All authors contributed to the study conception and design. Material preparation, data collection and analysis were performed by GYSCMC, JVF, CEFS and FOC. The first draft of the manuscript was written by BMVG, Lucas Meili and CEFS and all authors evaluated previous versions of the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest to disclose.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
De Farias Silva, C.E., Costa, G.Y.S.C.M., Ferro, J.V. et al. Application of machine learning to predict the yield of alginate lyase solid-state fermentation by Cunninghamella echinulata: artificial neural networks and support vector machine. Reac Kinet Mech Cat 135, 3155–3171 (2022). https://doi.org/10.1007/s11144-022-02293-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11144-022-02293-9