Abstract
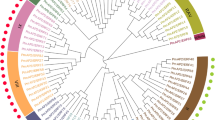
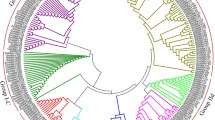
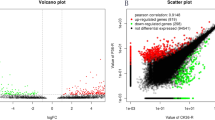
Masson pine (Pinus massoniana Lamb.) is an economically important conifer tree that can be widely used for timber, pulp, and resin production. However, the phosphate (Pi) deficiency in tropical and subtropical forest soils poses severe challenges for the productivity of masson pine. WRKY transcription factors (TFs) have been proven to play important roles in plant responses to biotic and abiotic stresses, including low Pi stress; however, little is known about their roles in masson pine. To understand the roles of P. massoniana WRKY (PmWRKY) in low Pi stress, 25 putative WRKY TFs with complete WRKY domain from transcriptome sequencing data were identified. Based on their conserved domains and zinc-finger motif, the P. massoniana WRKY were divided into three groups. Structural feature analysis shows that PmWRKY proteins contain WRKYGQK/GKK/GRK domains and a C2H2/C2HC-type zinc-finger structure. To putatively identify the function of PmWRKY members involved in low Pi stress, transcriptional profiles of 17 PmWRKYs in masson pine under different Pi stresses were systematically established using real-time quantitative RT-PCR analysis. Analysis demonstrates that the candidate PmWRKYs were involved in responses to Pi starvation—for example, PmWRKY11, 12, and 13 were upregulated both in P1 (Phosphorus content of 0.01 mM) and P2 (Phosphorus content of 0.06 mM) stresses. The available evidence conclusively sheds light on protein structures, evolutionary relationships, and expression patterns of WRKYs in response to low Pi stress of masson pine, which facilitates further functional identification and molecular breeding for the enhancement of low-phosphorous tolerance in this species.




Similar content being viewed by others
References
Atkinson NJ, Urwin PE (2012) The interaction of plant biotic and abiotic stresses: from genes to the field. J Exp Bot 63:3523–3543
Chen C, Chen Z (2000) Isolation and characterization of two pathogen- and salicylic acid-induced genes encoding WRKY DNA-binding proteins from tobacco. Plant Mol Biol 42:387–396
Chen L, Song Y, Li S, Zhang L, Zou C, Yu D (2012) The role of WRKY transcription factors in plant abiotic stresses. Biochim Biophys Acta 1819:120–128
Chen YF, Li LQ, Xu Q, Kong YH, Wang H, Wu WH (2009) The WRKY6 transcription factor modulates PHOSPHATE1 expression in response to low pi stress in Arabidopsis. Plant Cell 21:3554–3566
Ciolkowski I, Wanke D, Birkenbihl RP, Somsich I (2008) Studies on DNA-binding selectivity of WRKY transcription factors lend structural clues into WRKY-domain function. Plant Mol Biol 68:81–92
Cui Y, Xie H, Wang J (2005) Potential biomedical properties of Pinus massoniana bark extract. Phyto Res: In J Devo Pharma Toxic Eva Nat Pro Deriv 19:34–38
Dai X, Wang Y, Zhang W (2016) OsWRKY74, a WRKY transcription factor, modulates tolerance to phosphate starvation in rice. J Exp Bot 67:947–960
Eulgem T, Somssich IE (2007) Networks of WRKY transcription factors in defense signaling. Cur Opin Plant Biol 10:366–371
Eulgem T, Rushton PJ, Robatzek S, Somssich IE (2000) The WRKY superfamily of plant transcription factors. Trends Plant Sci 5:199–206
Fan F, Cui B, Zhang T, Qiao G, Wen X (2014) The temporal transcriptomic response of Pinus massoniana seedlings to phosphorus deficiency. PLoS One 9:e105068
Fan X, Guo Q, Xu P, Gong Y, Shu H, Yang Y, Ni W, Zhang X, Shen X (2015) Transcriptome-wide identification of salt-responsive members of the WRKY gene family in Gossypium aridum. PLoS One 10:e0126148
He H, Dong Q, Shao Y, Jiang H, Zhu S, Cheng B, Xiang Y (2012) Genome-wide survey and characterization of the WRKY gene family in Populus trichocarpa. Plant Cell Rep 31:1199–1217
Huang S, Gao Y, Liu J, Peng X, Niu X, Fei Z, Cao S, Liu Y (2012) Genome-wide analysis of WRKY transcription factors in Solanum lycopersicum. Mol Gen Genomics 287:495–513
Huang Y, Li M, Wu P, Xu Z, Que F, Wang F, Xiong A (2016) Members of WRKY Group III transcription factors are important in TYLCV defense signaling pathway in tomato (Solanum lycopersicum). BMC Genomics 17:788
Ichimura K, Mizoguchi T, Yoshida R, Yuasa T, Shinozaki K (2000) Various abiotic stresses rapidly activate Arabidopsis MAP kinases ATMPK4 and ATMPK6. Plant J 24:655–665
Ishiguro S, Nakamura K (1994) Characterization of a cDNA encoding a novel DNA-binding protein, SPF1, that recognizes SP8 sequences in the 5′ upstream regions of genes coding for sporamin and β-amylase from sweet potato. Mol Gen Genet MGG 244:563–571
Jian L, Jiang W, Ying Z, Yu H, Mao Z, Gu X, Huang S, Xie B (2011) Genome-wide analysis of WRKY gene family in Cucumis sativus. BMC Genomics 12:471
Jiang J, Ma S, Ye N, Jiang M, Cao J, Zhang J (2017) WRKY transcription factors in plant responses to stresses. J Integr Plant Bio 59:86–101
Jiang Y, Zeng B, Zhao H, Zhang M, Xie S, Lai J (2012) Genomewide transcription factor gene prediction and their expressional tissue-specificities in maize. J Int Plant Bio 54:616–630
Livak KJ, Schmittgen TD (2012) Analysis of relative gene expression data using real-time quantitative PCR and the 2 (-Delta Delta C(T)) Method. Methods 25:402–408
Nakashima K, Ito Y, Yamaguchi-Shinozaki K (2009) Transcriptional regulatory networks in response to abiotic stresses in Arabidopsis and grasses. Plant Physiol 149:88–95
Okay S, Derelli E, Unver T (2014) Transcriptome-wide identification of bread wheat WRKY transcription factors in response to drought stress. Mol Gen Genomics 289:765–781
Puranik S, Sahu PP, Mandal SN, Parida SK, Prasad M (2013) Comprehensive genome-wide survey, genomic constitution and expression profiling of the NAC transcription factor family in foxtail millet (Setaria italica L.). PLoS One 8:e64594
Rinerson CI, Rabara RC, Tripathi P, Shen Q, Rushton PJ (2015) The evolution of WRKY transcription factors. BMC Plant Biol 15:66
Rushton PJ, Macdonald H, Huttly AK, Lazarus CM, Hooley R (1995) Members of a new family of DNA-binding proteins bind to a conserved cis-element in the promoters of alpha-Amy2 genes. Plant Mol Biol 29:691–702
Rushton PJ, Somssich IE, Ringler P, Shen QJ (2010) WRKY transcription factors. Trends in Plant Sci 15:247–258
Shi W, Hao L, Li J, Liu D, Guo X, Li H (2014) The Gossypium hirsutum WRKY gene GhWRKY39-1 promotes pathogen infection defense responses and mediates salt stress tolerance in transgenic Nicotiana benthamiana. Plant Cell Rep 33:483–498
Su T, Xu Q, Zhang FC, Chen Y, Li LQ, Wu WH, Chen YF (2015) WRKY42 modulates phosphate homeostasis through regulating phosphate translocation and acquisition in Arabidopsis. Plant Physiol 167:1579–1591
Sun C, Palmqvist S, Olsson H, Borén M, Ahlandsberg S, Jansson C (2003) A novel WRKY transcription factor, SUSIBA2, participates in sugar signaling in barley by binding to the sugar-responsive elements of the iso1 promoter. Plant Cell 15:2076–2092
Ulker B, Somssich IE (2004) WRKY transcription factors: from DNA binding towards biological function. Curr Opin Plant Bio 7:491–498
van Verk MC, Pappaioannou D, Neeleman L, Bol JF, Linthorst HJ (2008) A novel WRKY transcription factor is required for induction of PR-1a gene expression by salicylic acid and bacterial elicitors. Plant Physiol 146:1983–1995
Wang J, Tao F, An F, Zou Y, Tian W, Chen X, Xu X, Hu X (2017) Wheat transcription factor TaWRKY70 is positively involved in high-temperature seedling-plant resistance to Puccinia striiformis f. sp. tritici. Mol Plant Patho 18:649–661
Wang M, Vannozzi A, Wang G, Liang Y, Tornielli GB, Zenoni S, Cavallini E, Pezzotti M, Cheng Z (2014) Genome and transcriptome analysis of the grapevine (Vitis vinifera L.) WRKY gene family. Hort Res 1:14016
Wu KL, Guo ZJ, Wang HH, Li J (2005) The WRKY family of transcription factors in rice and Arabidopsis and their origins. DNA Res 12:9–26
Wu Z, Li X, Liu Z, Li H, Wang Y, Zhuang J (2016) Transcriptome-wide identification of Camellia sinensis WRKY transcription factors in response to temperature stress. Mol Gen Genomics 291:255–269
Xie Z, Zhang ZL, Zou X, Huang J, Ruas P, Thompson D, Shen QJ (2005) Annotations and functional analyses of the rice WRKY gene superfamily reveal positive and negative regulators of abscisic acid signaling in aleurone cells. Plant Physiol 137:76–189
Xu H, Watanabe KA, Zhang L, Shen QJ (2016) WRKY transcription factor genes in wild rice Oryza nivara. DNA Res 23:311–323
Ye Q, Wang H, Su T, Wu WH, Chen YF (2018) The ubiquitin E3 ligase PRU1 regulates WRKY6 degradation to modulate phosphate homeostasis in response to low-pi stress in Arabidopsis. Plant Cell 30:1062–1076
Zhang L, Gu L, Ringler P, Smith S, Rushton PJ, Shen Q (2015) Three WRKY transcription factors additively repress abscisic acid and gibberellin signaling in aleurone cells. Plant Sci 236:214–222
Zhang Y, Wang L (2005) The WRKY transcription factor superfamily: its origin in eukaryotes and expansion in plants. BMC Evo Bio 5:1
Zhang Z, Xie Z, Zo X, Casaretto J, Ho T, Shen Q (2004) A rice WRKY gene encodes a transcriptional repressor of the gibberellin signaling pathway in aleurone cells. Plant Physiol 134:1500–1513
Acknowledgments
We sincerely thank Prof. Kevin Johnston for providing proofreading help.
Funding
This study was funded by the National Natural Science Foundation of China (31660185), the Natural Science Foundation of Guizhou Province, P. R. China (20161051), and the National Key Research and Development Project of China (2017YFD0600302, 2017YFD0600303).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interests.
Human and animal rights
The research described here did not involve human participants or animals.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Key Message
To understand P. massoniana WRKY (PmWRKY) roles in low Pi stress, we performed a transcriptome-wide identification of WRKY TFs in masson pine. Judging from their conserved domains and zinc-finger motif, they were diversely homologous to those from Arabidopsis thaliana and Oryza sativa, and we divided them into three groups. Structural feature analysis shows that the PmWRKY proteins contained WRKYGQK/GKK/GRK domains and a C2H2/C2HC-type zinc-finger structure. Transcriptional profiles of 17 PmWRKYs in masson pine upon different Pi stresses were systematically established using real-time quantitative RT-PCR analysis, and candidate PmWRKYs involved in responses to Pi-deficiency were identified.
Electronic supplementary material
ESM 1
(DOC 334642 kb)
Rights and permissions
About this article
Cite this article
Fan, F., Wang, Q., Li, H. et al. Transcriptome-Wide Identification and Expression Profiles of Masson Pine WRKY Transcription Factors in Response to Low Phosphorus Stress. Plant Mol Biol Rep 39, 1–9 (2021). https://doi.org/10.1007/s11105-020-01222-1
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11105-020-01222-1