Abstract
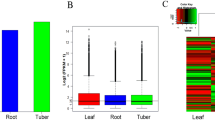
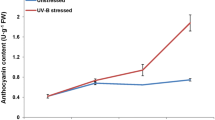
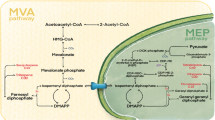
Hypericum perforatum is a traditional medicinal plant used for various purposes since ancient times because of the valuable secondary metabolites. The genomic information for H. perforatum is limited and the regulatory networks of secondary metabolism are still unknown. The naphthodianthrone hypericin is the metabolite of our main interest. It is a red-colored photodynamic pigment localized in dark glands of some Hypericum species. High-throughput sequencing technology, especially RNA-Seq, followed by de novo assembly and analysis of differential gene expression provides an important tool for functional genomics of non-model organisms. It represents an opportunity of insight into dynamic biological processes including those of secondary metabolism. Transcriptome analysis followed by the analysis of differential gene expression of H. perforatum leaf tissues containing and lacking dark glands was performed to identify the genes involved in hypericin biosynthesis and to profile expression patterns in greater detail. A total of 18.53 G of cleaned read nucleotides were generated and assembled into 139,959 contigs with N50 of 1801 bp. Among them, 66,817 (47.74 %) contigs were annotated. Differentially expressed genes were discovered by comparison of dark glands with adjacent leaf tissue and contrasting inner part leaf tissue without dark glands. A total of 799 upregulated genes were found in the tissues containing dark glands and 263 enzymes were identified, including candidate genes of hypericin biosynthesis, especially the genes coding for polyketide synthases and those involved in defense reactions. This study determined candidate genes involved in hypericin biosynthesis providing a valuable source for perspective metabolic engineering of bioactive substances.









Similar content being viewed by others
References
Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, Sherlock G (2000) Consortium GO: Gene Ontology: tool for the unification of biology. Nat Genet 25:25–29
Austin MB, Noel JP (2003) The chalcone synthase superfamily of type III polyketide synthases. Nat Prod 20:79–110
Bais HP, Vepachedu R, Lawrence CB, Stermitz FR, Vivanco JM (2003) Molecular and biochemical characterization of an enzyme responsible for the formation of hypericin in St. John’s Wort (Hypericum perforatum L.). J Biol Chem 278(34):32413–22
Banks HJ, Cameron DW, Raverty WD (1976) Chemistry of the coccoidea. II Condensed polycyclic pigments from two Australian pseudococcids (Hemiptera). Aust J Chem 29:1509
Chen Z, Fujii I, Ebizuka Y, Sankawa U (1995) Purification and characterization of emodinanthrone oxygenase from Aspergillus terreus. Phytochemistry 38:299–305
Ciccarelli D, Andreucci AC, Pagni AM (2001) Translucent glands and secretory canals in Hypericum Perforatum L. (Hypericaceae): morphological, anatomical and histochemical studies during the course of ontogenesis. Ann Bot-London 88:637–44
Conesa A, Götz S (2008) Blast2GO: a comprehensive suite for functional analysis in plant genomics. Int J Plant Genomics 2008:619832
Crockett SL, Robson NKB (2011) Taxonomy and chemotaxonomy of the genus Hypericum. MAPSB 5:1–13
Curtis JD, Lersten NR (1990) Internal secretory structures in Hypericum (Clusiaceae): H. perforatum L. and H. balearicum L. New Phytol 114:571–580
Fu L, Niu B, Zhu Z, Wu S, Li W (2012) CD-HIT: accelerated for clustering the next generation sequencing data. Bioinformatics 28:3150–3152
Galla G, Vogel H, Sharbel TF, Barcaccia G (2015) De novo sequencing of the Hypericum perforatum L. flower transcriptome to identify potential genes that are related to plant reproduction sensu lato. BMC Genomics 16:254
Gamborg OL, Miller RA, Ojima K (1968) Nutrient requirements of suspension cultures of soybean root cells. Exp Cell Res 50:151–158
Gill M, Gimenez A (1991) Austrovenetin, the principal pigment of the toadstool Dermocybe austroveneta. Phytochemistry 30:951–955
Gill M, Gimenez A, McKenzie RW (1988) Pigments of fungi, part 8. Bianthraquinones from Dermocybe austroveneta. J Nat Prod 51:1251–1256
Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, Adiconis X, Fan L, Raychowdhury R, Zeng Q, Chen Z, Mauceli E, Hacohen N, Gnirke A, Rhind N, di Palma F, Birren BW, Nusbaum C, Lindblad-Toh K, Friedman N, Regev A (2011) Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol 29:644–652
He M, Wang Y, Hua W, Zhang Y, Wang Z (2012) De novo sequencing of Hypericum Perforatum transcriptome to identify potential genes involved in the biosynthesis of active metabolites. PLoS One 7, e42081
Huang N, Rizshsky L, Hauck C, Nikolau BJ, Murphy PA, Birt DF (2011) Identification of anti-inflammatory constituents in Hypericum Perforatum and Hypericum Gentianoides extracts using RAW 264.7 mouse macrophages. Phytochemistry 72:2015–23
Huang HL, Wang ZH, Chen SL (2014) Hypericin: chemical synthesis and biosynthesis. CJNM 12:81–88
Kanehisa M, Goto S, Sato Y, Kawashima M, Furumichi M, Tanabe M (2014) Data, information, knowledge and principle: back to metabolism in KEGG. Nucleic Acids Res 42:199–205
Karioti A, Bilia AR (2010) Hypericins as potential leads for new therapeutics. Int J Mol Sci 11:562–94
Karppinen K, Hohtola A (2008) Molecular cloning and tissue-specific expression of two cDNAs encoding polyketide synthases from Hypericum perforatum. J Plant Physiol 165:1079–1086
Karppinen K, Hokkanen J, Mattila S, Neubauer P, Hohtola A (2008) Octaketide-producing type III polyketide synthase from Hypericum perforatum is expressed in dark glands accumulating hypericin. Febs J 275:4329–4342
Košuth J, Katkovčíková Z, Olexová P, Čellárová E (2007) Expression of the hyp-1 gene in early stages of development of Hypericum perforatum L. Plant Cell Rep 26:211–217
Košuth J, Smelcerovic A, Borsch T, Zuehlke S, Karppinen K, Spiteller M, Hohtola A, Čellárová E (2011) The hyp-1 gene is not a limiting factor for hypericin biosynthesis in the genus Hypericum. Funct Plan Biol 38:35–43
Košuth J, Hrehorová D, Jaskolski M, Čellárová E (2013) Stress-induced expression and structure of the putative gene hyp-1 for hypericin biosynthesis. Plant Cell Tiss Orga Cult 114:207–216
Kusari S, Lamshöft M, Zühlke S, Spiteller M (2008) An endophytic fungus from that produces hypericin. J Nal Prod 71:159–162
Kusari S, Selahaddin SS, Nigutová K, Čellárová E, Spiteller M (2015) Spatial chemo-profiling of hypericin and related phytochemicals in Hypericum species using MALDI-HRMS imaging. Anal Bioanal Chem 407:4779–4791
Langmead B, Trapnell C, Pop M, Salzberg SL (2009) Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10:R25
Li B, Dewey CN (2011) RSEM: accurate transcript quantification from RNA Seq data with or without a reference genome. BMC bioinformatics 12:323
Linde K, Knuppel L (2005) Large-scale observational studies of hypericum extracts in patients with depressive disorders—a systematic review. Phytomedicine 12:148–157
Liu B, Falkenstein-Paul H, Schmidt W (2003) Beerhues L. Benzophenone synthase and chalcone synthase from Hypericum androsaemum cell cultures: cDNA cloning, functional expression, and site-directed mutagenesis of two polyketide synthases. Plant J 34:847–855
Love MI, Huber W, Anders S (2014) Moderated estimation of fold change and dispersion for RNA-Seq data with DESeq2. Genome Biol 15:550
Martin M (2010) Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal 17:10–12
Michalska K, Fernandes H, Sikorski M, Jaskolski M (2010) Crystal structure of Hyp-1, a St. John´s wort protein implicated in the biosynthesis of hypericin. J Struct Biol 169:161–171
Murashige T, Skoog F (1962) A revised medium for rapid growth and bioassays with tobacco tissue cultures. Plant Physiolog 15:473–497
Myhre S, Tveit H, Mollestad T, Laegreid A (2006) Additional gene ontology structure for improved biological reasoning. Bioinformatics 22:2020–2027
Onelli E, Rivetta A, Giorgi A, Bignami M, Cocucci M, Patrignani G (2002) Ultrastructural studies on the developing secretory nodules of Hypericum perforatum. New Phytol 197:92–102
Pillai PP, Nair AR (2014) Hypericin biosynthesis in Hypericum hookerianum Wight and Arn: investigation on biochemical pathways using metabolite inhibitors and suppression substractive hybridization. C R Biologies 337:571–580
Robson NKB (2003) Hypericum botany. In: Ernst E (ed) Hypericum—the genus Hypericum. Taylor and Francis, London, New York, pp 1–22
Sirvent T, Gibson D (2002) Induction of hypericin and hyperforin in Hypericum perforatum L. in response to biotic and chemici elicitors. Physiol Mol Plant P60:311–320
Strickler SR, Bombarely A, Mueller LA (2012) Designing a transcriptome next-generation sequencing project for a nonmodel plant species. Am J Bot 99:257–66
Walker EB, Lee T, Song PS (1979) Spectroscopic characterization of the Stentor photoreceptor. Biochim Biophys Acta 587:129–144
Wolkenstein K, Gross JH, Falk H, Schöler HF (2006) Preservation of hypericin and related polycyclic quinone pigments in fossil crinoids. Proc Biol Sci 273:451–56
Xiao M, Zhang Z, Chen X, Lee E, Barber CJS, Chakrabarty R, Desgagné-Penix I, Haslam TM, Kim Y, Liu E, MacNevin G, Masada-Atsumi S, Reed D, Stout JM, Zerbe P, Zhang Y, Bohlmann J, Covello PS, Luca V, Page JE, Ro D, Martin VJJ, Facchini PJ, Sensen CW (2013) Transcriptome analysis based on next-generation sequencing of non-model plants producing specialized metabolites of biotechnological interest. J Biotechnol 166:122–134
Ye J, Fang L, Zheng H, Zhang Y, Chen J, Zhang Z, Wang J, Li S, Li R, Bolund L, Wang J (2006) WEGO: a web tool for plotting GO annotations. Nucleic Acids Res 34:293–297
Zdobnow EM, Apweiler R (2001) InterProScan—an integration platform for the signature-recognition methods in InterPro. Bioinformatics 17:847–848
Zhao J, Davis LC, Verpoorte R (2005) Elicitor signal transduction leading to production of plant secondary metabolites. Biotechnol Adv 23:283–333
Zobayed SM, Afreen F, Goto E, Kozai T (2006) Plant-environment interactions: accumulation of hypericin in dark glands of Hypericum perforatum. Ann Bot-London 98:793–804
Acknowledgments
This research was supported by the Slovak Research and Development Agency APVV-14-0154, the Scientific Grant Agency of Slovak Republic VEGA 1/0090/15, and the grant project SOFOS-knowledge and skill development of staff, students of P. J. Šafárik University in Košice (contract number: 003/2013/1.2/OPV, ITMS code: 26110230088), funded by the European Social Fund through the Operational Program Education and KVARK (ITMS code: 26110230084). We thank Peter Pisarčík, Maroš Andrejko, Tomáš Horváth, and Gabriel Semanišin from Institute of Computer Sciences, Faculty of Science, Pavol Jozef Šafárik University, in Košice and Libuse Brachova from Roy J. Carver Department of Biochemistry, Biophysics, and Molecular Biology, Iowa State University, for support.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1: Table S1
Upregulated differential expressed genes of dark glands with adjacent leaf tissue of H. perforatum annotated as oxidoreductases. (DOCX 32 kb)
ESM 2: Table S2
Upregulated differential expressed genes of dark glands with adjacent leaf tissue of H. perforatum annotated as transferases. (DOCX 38 kb)
ESM 3: Table S3
Upregulated differential expressed genes of dark glands with adjacent leaf tissue of H. perforatum annotated as hydrolases. (DOCX 37 kb)
ESM 4: Table S4
Upregulated differential expressed genes of dark glands with adjacent leaf tissue of H. perforatum annotated as lyases. (DOCX 15 kb)
ESM 5: Table S5
Upregulated differential expressed genes of dark glands with adjacent leaf tissue of H. perforatum annotated as isomerases. (DOCX 12 kb)
ESM 6: Table S6
Upregulated differential expressed genes of dark glands with adjacent leaf tissue of H. perforatum annotated as ligases. (DOCX 14 kb)
Rights and permissions
About this article
Cite this article
Soták, M., Czeranková, O., Klein, D. et al. Differentially Expressed Genes in Hypericin-Containing Hypericum perforatum Leaf Tissues as Revealed by De Novo Assembly of RNA-Seq. Plant Mol Biol Rep 34, 1027–1041 (2016). https://doi.org/10.1007/s11105-016-0982-2
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11105-016-0982-2