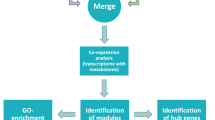
Abstract
Rauvolfia serpentina has been known to produce therapeutically important indole alkaloids used in treatment of various diseases. Despite its medicinal importance, complete understanding of its secondary metabolism is challenging due to complex interplay among various transcription factors (TFs) and genes. However, weighted co-expression analysis of transcriptome along with integration of metabolomics data has proficiency to elucidate topological properties of complex regulatory interactions in secondary metabolism. We aimed to implement an integrative strategy using “-omics” data to identify TFs of “unknown function” and exemplify their role in regulation of valuable metabolites as well as metabolic traits. A total of 69 TFs were identified through significant thresholds and removal of false positives based on cis-regulatory motif analysis. Network-biology inspired analysis of co-expression network lead to generation of four statistically significant and biologically robust modules. Similar to known regulatory roles of WRKY and AP2-EREBP TF families in Catharanthus roseus, this study presented them to regulate synthesis of alkaloids in R. serpentina as well. Moreover, TFs in module 4 were observed to be regulating connecting steps between primary and secondary metabolic pathways in the synthesis of terpenoid indole alkaloids. Integration of metabolomics data further highlight the significance of module 1 since it was statistically predicted to be involved in synthesis of specialized metabolites, and associated genes may physically clustered on genome. Importantly, putative TFs in module 1 may modulate the major indole alkaloids synthesis in response to various environmental stimuli. The methodology implemented herein may provide a better reference to identify and explore functions of transcriptional regulators.








Similar content being viewed by others
Abbreviations
- TFs:
-
Transcription factors
- PCC:
-
Pearson correlation coefficient
- MPGR:
-
Medicinal plant genomics resource
- TAIR10:
-
The Arabidopsis Information Resource 10
- PlnTFDB:
-
Plant Transcription Factor Database
- ND:
-
Network density
- AGRIS:
-
Arabidopsis gene regulatory information server
- TFBS:
-
Transcription factor binding sites
- MCL:
-
Markov cluster
- GO:
-
Gene ontology
- KEGG:
-
Kyoto Encyclopedia of Genes and Genomes
- PMR:
-
Plant and microbial metabolomics resource
- TIA:
-
Terpenoid indole alkaloid
References
Aharoni A, Galili G (2011) Metabolic engineering of the plant primary-secondary metabolism interface. Curr Opin Biotechnol 22:239–244. doi:10.1016/j.copbio.2010.11.004
Albert R (2005) Scale-free networks in cell biology. J Cell Sci 118:4947–57. doi:10.1242/jcs.02714
Alon U (2006) An introduction to systems biology: design principles of biological circuits, 1st edn. 320
Altschul SF, Madden TL, Schäffer AA et al (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402. doi:10.1093/nar/25.17.3389
Aniszewski T (2007) Alkaloids-secrets of life: alkaloid chemistry, biological significance, applications and ecological role, 1st edn. 334
Aoki K, Ogata Y, Shibata D (2007) Approaches for extracting practical information from gene co-expression networks in plant biology. Plant Cell Physiol 48:381–390. doi:10.1093/pcp/pcm013
Azmi M, Qureshi A (2012) Methanolic root extract of Rauwolfia serpentina improves the glucose tolerance in wister mice. J Food Drug Anal 20:484–488. doi:10.6227/jfda.2012200208
Baroux C, Pien S, Grossniklaus U (2007) Chromatin modification and remodeling during early seed development. Curr Opin Genet Dev 17:473–479. doi:10.1016/j.gde.2007.09.004
Barrero RA, Chapman B, Yang Y et al (2011) De novo assembly of Euphorbia fischeriana root transcriptome identifies prostratin pathway related genes. BMC Genomics 12:600. doi:10.1186/1471-2164-12-600
Bazika V (1969) Ajmaline, an alkaloid of Rauwolfia serpentina, in the treatment of cardiac arrhythmias. Vnitr Lek 15:662–670
Beljanski M, Beljanski MS (1982) Selective inhibition of in vitro synthesis of cancer DNA by alkaloids of beta-carboline class. Exp Cell Biol 50:79–87
Benjamini Y, Hochberg Y (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Ser B 57:289–300. doi:10.2307/2346101
Bindea G, Mlecnik B, Hackl H et al (2009) ClueGO: a Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics 25:1091–1093. doi:10.1093/bioinformatics/btp101
Borevitz JO (2000) Activation tagging identifies a conserved MYB regulator of phenylpropanoid biosynthesis. Plant Cell Online 12:2383–2394. doi:10.1105/tpc.12.12.2383
Chae L, Kim T, Nilo-Poyanco R, Rhee SY (2014) Genomic signatures of specialized metabolism in plants. Science (80) 344:510–513. doi: 10.1126/science.1252076
Chang K, Chen M, Zeng L et al (2013) Abscisic acid enhanced ajmalicine biosynthesis in hairy roots of Rauvolfia verticillata by upregulating expression of the MEP pathway genes. Russ J Plant Physiol 61:136–140. doi:10.1134/S102144371401004X
Chien C-H, Chiang-Hsieh Y-F, Tsou A-P et al (2014) Large-scale investigation of human TF-miRNA relations based on coexpression profiles. Biomed Res Int. doi:10.1155/2014/623078
Childs KL, Davidson RM, Buell CR (2011) Gene coexpression network analysis as a source of functional annotation for rice genes. PLoS One 6:e22196. doi:10.1371/journal.pone.0022196
Chou WM, Kutchan TM (1998) Enzymatic oxidations in the biosynthesis of complex alkaloids. Plant J 15:289–300. doi:10.1046/j.1365-313X.1998.00220.x
Chou W-C, Cheng A-L, Brotto M, Chuang C-Y (2014) Visual gene-network analysis reveals the cancer gene co-expression in human endometrial cancer. BMC Genomics 15:300. doi:10.1186/1471-2164-15-300
Chu HY, Wegel E, Osbourn A (2011) From hormones to secondary metabolism: the emergence of metabolic gene clusters in plants. Plant J 66:66–79. doi:10.1111/j.1365-313X.2011.04503.x
Csardi G, Nepusz T (2006) The igraph software package for complex network research. InterJournal Complex Sy:1695
D’Auria JC, Gershenzon J (2005) The secondary metabolism of Arabidopsis thaliana: growing like a weed. Curr Opin Plant Biol 8:308–316. doi:10.1016/j.pbi.2005.03.012
Davuluri RV, Sun H, Palaniswamy SK et al (2003) AGRIS:Arabidopsis gene regulatory information server, an information resource of Arabidopsis cis-regulatory elements and transcription factors. BMC Bioinf 4:25. doi:10.1186/1471-2105-4-25
De Luca V, St Pierre B (2000) The cell and developmental biology of alkaloid biosynthesis. Trends Plant Sci 5:168–173. doi: 10.1016/S1360-1385(00)01575-2
De Vega-Bartol JJ, Simões M, Lorenz WW et al (2013) Transcriptomic analysis highlights epigenetic and transcriptional regulation during zygotic embryo development of Pinus pinaster. BMC Plant Biol. doi:10.1186/1471-2229-13-123
Dey A, De J (2011) Ethnobotanical aspects of Rauvolfia serpentina (L). Benth. ex Kurz. in India, Nepal and Bangladesh. J Med Plants Res 5:144–150
Dimmer EC, Huntley RP, Barrell DG, et al (2008) The Gene Ontology - providing a functional role in proteomic studies. Proteomics 8:n/a. doi: 10.1002/pmic.200800002
Du Z, Zhou X, Ling Y et al (2010) agriGO: a GO analysis toolkit for the agricultural community. Nucleic Acids Res 38:W64–W70. doi:10.1093/nar/gkq310
Dubey VS, Bhalla R, Luthra R (2003) An overview of the non-mevalonate pathway for terpenoid biosynthesis in plants. J Biosci 28:637–646. doi:10.1007/BF02703339
Ehrlich PR, Raven PH (1964) Butterflies and plants: a study in coevolution. Evol (N Y) 18:586–608
Eichinger D (1999) Analysis of metabolic pathways via quantitative prediction of isotope labeling patterns: a retrobiosynthetic 13C NMR study on the monoterpene loganin. Phytochemistry 51:223–236. doi:10.1016/S0031-9422(98)00733-X
Eisen MB, Spellman PT, Brown PO, Botstein D (1998) Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci 95:14863–14868. doi:10.1073/pnas.95.25.14863
El-Sayed M, Verpoorte R (2007) Catharanthus terpenoid indole alkaloids: biosynthesis and regulation. Phytochem Rev 6:277–305. doi:10.1007/s11101-006-9047-8
Emmert-streib F, Dehmer M (2008) Robustness in scale-free networks: comparing directed and undirected networks. Int J Mod Phys C 19:717–726. doi:10.1142/S0129183108012510
Ershov YV, Gantt RR, Cunningham FX, Gantt E (2002) Isoprenoid biosynthesis in Synechocystis sp. strain PCC6803 is stimulated by compounds of the pentose phosphate cycle but Not by pyruvate or deoxyxylulose-5-phosphate. J Bacteriol 184:5045–5051. doi:10.1128/JB.184.18.5045-5051.2002
Facchini PJ, De Luca V (2008) Opium poppy and Madagascar periwinkle: model non-model systems to investigate alkaloid biosynthesis in plants. Plant J 54:763–784. doi:10.1111/j.1365-313X.2008.03438.x
Fauteux F, Strömvik MV (2009) Seed storage protein gene promoters contain conserved DNA motifs in Brassicaceae, Fabaceae and Poaceae. BMC Pant Biol 9:126. doi:10.1186/1471-2229-9-126
Fernandez JA, de Luca V (1994) Ubiquitin-mediated degradation of tryptophan decarboxylase from Catharanthus roseus. Phytochemistry 36:1123–1128. doi:10.1016/S0031-9422(00)89624-7
Flügge U-I, Gao W (2005) Transport of isoprenoid intermediates across chloroplast envelope membranes. Plant Biol 7:91–97. doi:10.1055/s-2004-830446
Fraenkel GS (1959) The Raison d’Etre of Secondary Plant Substances: These odd chemicals arose as a means of protecting plants from insects and now guide insects to food. Science (80- ) 129:1466–1470. doi: 10.1126/science.129.3361.1466
Frey M, Huber K, Park WJ et al (2003) A 2-oxoglutarate-dependent dioxygenase is integrated in DIMBOA-biosynthesis. Phytochemistry 62:371–376. doi:10.1016/S0031-9422(02)00556-3
Fukushima A, Nishizawa T, Hayakumo M et al (2012) Exploring tomato gene functions based on coexpression modules using graph clustering and differential coexpression approaches. Plant Physiol 158:1487–1502. doi:10.1104/pp. 111.188367
Glenn WS, Runguphan W, O’Connor SE (2013) Recent progress in the metabolic engineering of alkaloids in plant systems. Curr Opin Biotechnol 24:354–365. doi:10.1016/j.copbio.2012.08.003
Gongora-Castillo E, Childs KL, Fedewa G et al (2012) Development of transcriptomic resources for interrogating the biosynthesis of monoterpene indole alkaloids in medicinal plant species. PLoS One 7:e52506. doi:10.1371/journal.pone.0052506
Grafi G, Ohad N (2013) Epigenetic Memory and Control in Plants. 270
Hartwell LH, Hopfield JJ, Leibler S, Murray AW (1999) From molecular to modular cell biology. Nature 402:C47–C52. doi:10.1038/35011540
Haynes BC, Maier EJ, Kramer MH et al (2013) Mapping functional transcription factor networks from gene expression data. Genome Res 23:1319–1328. doi:10.1101/gr.150904.112
He K, Guo A-Y, Gao G et al (2010) Computational identification of plant transcription factors and the construction of the PlantTFDB database. Methods Mol Biol 674:351–368. doi:10.1007/978-1-60761-854-6_21
Herbert RB (2003) The biosynthesis of plant alkaloids and nitrogenous microbial metabolites. Nat Prod Rep 20:494–508. doi:10.1039/b006522f
Herrmann KM, Weaver LM (1999) The shikimate pathway. Annu Rev Plant Physiol Plant Mol Biol 50:473–503. doi:10.1146/annurev.arplant.50.1.473
Hirai MY, Klein M, Fujikawa Y et al (2005) Elucidation of gene-to-gene and metabolite-to-gene networks in Arabidopsis by integration of metabolomics and transcriptomics. J Biol Chem 280:25590–255595. doi:10.1074/jbc.M502332200
Hu J, Wang D, Li J et al (2014) Genome-wide identification of transcription factors and transcription-factor binding sites in oleaginous microalgae Nannochloropsis. Sci Rep 4:5454. doi:10.1038/srep05454
Hutcheon DE (1971) The pharmacological basis of therapeutics, 4th Edition. Louis S. Goodman and Alfred Gilman. J Clin Pharmacol New Drugs 11:388. doi:10.1177/009127007101100511
Ihmels J, Levy R, Barkai N (2004) Principles of transcriptional control in the metabolic network of Saccharomyces cerevisiae. Nat Biotechnol 22:86–92. doi:10.1038/nbt918
Jakoby M, Weisshaar B, Dröge-Laser W et al (2002) bZIP transcription factors in Arabidopsis. Trends Plant Sci 7:106–111. doi:10.1016/S1360-1385(01)02223-3
Jerie P (2007) Milestones of cardiovascular therapy. IV. Reserpine. Cas Lek Cesk 146:573–577
Jha A, Shankar R (2014) MiRNAting control of DNA methylation. J Biosci 39:365–380. doi:10.1007/s12038-014-9437-9
Jha A, Mehra M, Shankar R (2011) The regulatory epicenter of miRNAs. J Biosci 36:621–638. doi:10.1093/nar/gkj143
Jiao Y, Lau OS, Deng XW (2007) Light-regulated transcriptional networks in higher plants. Nat Rev Genet 8:217–230. doi:10.1038/nrg2049
Johnson S, Torres JJ, Marro J, Muñoz MA (2010) Entropic origin of disassortativity in complex networks. Phys Rev Lett 104:108702. doi:10.1103/PhysRevLett.104.108702
Kinsella RJ, Kähäri A, Haider S, et al (2011) Ensembl BioMarts: a hub for data retrieval across taxonomic space. Database (Oxford) 2011:bar030. doi: 10.1093/database/bar030
Koch MA, Weisshaar B, Kroymann J et al (2001) Comparative genomics and regulatory evolution: conservation and function of the Chs and Apetala3 promoters. Mol Biol Evol 18:1882–1891
Köppel C, Wagemann A, Martens F (1989) Pharmacokinetics and antiarrhythmic efficacy of intravenous ajmaline in ventricular arrhythmia of acute onset. Eur J Drug Metab Pharmacokinet 14:161–167. doi:10.1007/BF03190857
Kutchan TM (2005) A role for intra- and intercellular translocation in natural product biosynthesis. Curr Opin Plant Biol 8:292–300. doi:10.1016/j.pbi.2005.03.009
Lee WP, Tzou W-S (2009) Computational methods for discovering gene networks from expression data. Brief Bioinform 10:408–423. doi:10.1093/bib/bbp028
Lelek I, Furedi Szabo M (1961) On the antiallergic effect of the reserpine alkaloid of Rauwolfia serpentina. Allerg Asthma (Leipz) 7:142–144
Leménager D, Ouelhazi L, Mahroug S et al (2005) Purification, molecular cloning, and cell-specific gene expression of the alkaloid-accumulation associated protein CrPS in Catharanthus roseus. J Exp Bot 56:1221–1228. doi:10.1093/jxb/eri116
Li CY, Leopold AL, Sander GW et al (2013) The ORCA2 transcription factor plays a key role in regulation of the terpenoid indole alkaloid pathway. Plant Biol 13:155. doi:10.1186/1471-2229-13-155
Liang H, Li W-H (2007) MicroRNA regulation of human protein protein interaction network. RNA 13:1402–1408. doi:10.1261/rna.634607
Locket S (1955) Oral preparations of Rauwolfia serpentina in treatment of essential hypertension. Br Med J 1:809–816. doi:10.1136/bmj.1.4917.809
Ma S, Shah S, Bohnert HJ et al (2013) Incorporating motif analysis into gene co-expression networks reveals novel modular expression pattern and new signaling pathways. PLoS Genet 9:e1003840. doi:10.1371/journal.pgen.1003840
Mao L, Van Hemert JL, Dash S, Dickerson JA (2009) Arabidopsis gene co-expression network and its functional modules. BMC Bioinf 10:346. doi:10.1186/1471-2105-10-346
Matys V, Kel-Margoulis OV, Fricke E et al (2006) TRANSFAC and its module TRANSCompel: transcriptional gene regulation in eukaryotes. Nucleic Acids Res 34:D108–D110. doi:10.1093/nar/gkj143
Matzke M, Kanno T, Huettel B et al (2007) Targets of RNA-directed DNA methylation. Curr Opin Plant Biol 10:512–519. doi:10.1016/j.pbi.2007.06.007
McCluskey A, Lalkhen AG (2007) Statistics IV: interpreting the results of statistical tests. Contin Educ Anaesthesia, Crit Care Pain 7:208–212. doi:10.1093/bjaceaccp/mkm042
Memelink J, Gantet P (2007) Transcription factors involved in terpenoid indole alkaloid biosynthesis in Catharanthus roseus. Phytochem Rev 6:353–362. doi:10.1007/s11101-006-9051-z
Meyer E, Aspinwall MJ, Lowry DB et al (2014) Integrating transcriptional, metabolomic, and physiological responses to drought stress and recovery in switchgrass (Panicum virgatum L.). BMC Genomics 15:527. doi:10.1186/1471-2164-15-527
Misra A, Sriram G (2013) Network component analysis provides quantitative insights on an Arabidopsis transcription factor-gene regulatory network. BMC Syst Biol 7:126. doi:10.1186/1752-0509-7-126
Mitsuda N, Ohme-Takagi M (2009) Functional analysis of transcription factors in Arabidopsis. Plant Cell Physiol 50:1232–1248. doi:10.1093/pcp/pcp075
Mol J, Grotewold E, Koes R (1998) How genes paint flowers and seeds. Trends Plant Sci 3:212–217. doi:10.1016/S1360-1385(98)01242-4
Morris JH, Apeltsin L, Newman AM et al (2011) clusterMaker: a multi-algorithm clustering plugin for Cytoscape. BMC Bioinf 12:436. doi:10.1186/1471-2105-12-436
Naika M, Shameer K, Sowdhamini R (2013) Comparative analyses of stress-responsive genes in Arabidopsis thaliana: insight from genomic data mining, functional enrichment, pathway analysis and phenomics. Mol Biosyst 9:1888–1908. doi:10.1039/C3MB70072K
Nammi S, Boini KM, Koppula S, Sreemantula S (2005) Reserpine-induced central effects: pharmacological evidence for the lack of central effects of reserpine methiodide. Can J Physiol Pharmacol 83:509–515. doi:10.1139/y05-039
Nützmann H-W, Osbourn A (2014) Gene clustering in plant specialized metabolism. Curr Opin Biotechnol 26:91–99. doi:10.1016/j.copbio.2013.10.009
O’Connor SE, Maresh JJ (2006) Chemistry and biology of monoterpene indole alkaloid biosynthesis. Nat Prod Rep 23:532–547. doi:10.1039/B512615K
Oudin A, Courtois M, Rideau M, Clastre M (2007) The iridoid pathway in Catharanthus roseus alkaloid biosynthesis. Phytochem Rev 6:259–276. doi:10.1007/s11101-006-9054-9
Pani A, Mahapatra RK (2013) Computational identification of microRNAs and their targets in Catharanthus roseus expressed sequence tags. Genomics Data 1:2–6. doi:10.1016/j.gdata.2013.06.001
Pathania S, Randhawa V, Bagler G (2013) Prospecting for novel plant-derived molecules of Rauvolfia serpentina as inhibitors of Aldose Reductase, a potent drug target for diabetes and its complications. PLoS One 8:e61327. doi:10.1371/journal.pone.0061327
Paul A, Jha A, Bhardwaj S et al (2014) RNA-seq-mediated transcriptome analysis of actively growing and winter dormant shoots identifies non-deciduous habit of evergreen tree tea during winters. Sci Rep 4:5932. doi:10.1038/srep05932
Perianez-Rodriguez J, Manzano C, Moreno-Risueno MA (2014) Post-embryonic organogenesis and plant regeneration from tissues: two sides of the same coin. Front Plant Sci. doi:10.3389/fpls.2014.00219
Ramachandran S, Hiratsuka K, Chua NH (1994) Transcription factors in plant growth and development. Curr Opin Genet Dev 4:642–646. doi:10.1016/0959-437X(94)90129-Q
Raval A, Ray A (2013) Introduction to biological networks, 1st edn. 335.
Riaño-Pachón DM, Ruzicic S, Dreyer I, Mueller-Roeber B (2007) PlnTFDB: an integrative plant transcription factor database. BMC Bioinf 8:42. doi:10.1186/1471-2105-8-42
Ríos G, Leida C, Conejero A, Badenes ML (2014) Epigenetic regulation of bud dormancy events in perennial plants. Front Plant Sci 5:247. doi:10.3389/fpls.2014.00247
Ruan J, Dean AK, Zhang W (2010) A general co-expression network-based approach to gene expression analysis: comparison and applications. BMC Syst Biol. doi:10.1186/1752-0509-4-8
Ruppert M, Woll J, Giritch A et al (2005) Functional expression of an ajmaline pathway-specific esterase from Rauvolfia in a novel plant-virus expression system. Planta 222:888–98. doi:10.1007/s00425-005-0031-0
Samuni-Blank M, Izhaki I, Dearing MD et al (2012) Intraspecific directed deterrence by the mustard oil bomb in a desert plant. Curr Biol 22:1218–1220. doi:10.1016/j.cub.2012.04.051
Schluttenhofer C, Pattanaik S, Patra B, Yuan L (2014) Analyses of Catharanthus roseus and Arabidopsis thaliana WRKY transcription factors reveal involvement in jasmonate signaling. BMC Genomics 15:502. doi:10.1186/1471-2164-15-502
Schmid J, Amrhein N (1995) Molecular organization of the shikimate pathway in higher plants. Phytochemistry 39:737–749. doi:10.1016/0031-9422(94)00962-S
Schmid R, Blaxter ML (2008) annot8r: GO, EC and KEGG annotation of EST datasets. BMC Bioinf 9:180. doi:10.1186/1471-2105-9-180
Schuster G, Gruissem W (1991) Chloroplast mRNA 3’ end processing requires a nuclear-encoded RNA-binding protein. EMBO J 10:1493–1502
Seemann M, Tse Sum Bui B, Wolff M et al (2006) Isoprenoid biosynthesis in plant chloroplasts via the MEP pathway: direct thylakoid/ferredoxin-dependent photoreduction of GcpE/IspG. FEBS Lett 580:1547–1552. doi:10.1016/j.febslet.2006.01.082
Shannon P, Markiel A, Ozier O et al (2003) Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 13:2498–2504. doi:10.1101/gr.1239303
Shimizu M, Fujimoto R, Ying H et al (2014) Identification of candidate genes for Fusarium yellows resistance in Chinese cabbage by differential expression analysis. Plant Mol Biol 85:247–57. doi:10.1007/s11103-014-0182-0
Shoji T, Yamada Y, Hashimoto T (2000) Jasmonate induction of putrescine N-methyltransferase genes in the root of Nicotiana sylvestris. Plant Cell Physiol 41:831–839
Shukla AK, Shasany AK, Gupta MM, Khanuja SPS (2006) Transcriptome analysis in Catharanthus roseus leaves and roots for comparative terpenoid indole alkaloid profiles. J Exp Bot 57:3921–3932. doi:10.1093/jxb/erl146
Siberil Y (2002) Cloning of a cDNA encoding an E2 ubiquitin-conjugating enzyme from Catharanthus roseus: expression analysis in plant organs and in response to hormones in cell suspensions. J Exp Bot 53:149–150. doi:10.1093/jexbot/53.366.149
Sim J, Wright CC (2005) The kappa statistic in reliability studies: use, interpretation, and sample size requirements. Phys Ther 85:257–268
Simes RJ (1986) An improved Bonferroni procedure for multiple tests of significance. Biometrika 73:751–754. doi:10.1093/biomet/73.3.751
Singh K, Foley RC, Oñate-Sánchez L (2002) Transcription factors in plant defense and stress responses. Curr Opin Plant Biol 5:430–436. doi:10.1016/S1369-5266(02)00289-3
Singh DK, Srivastava B, Sahu A (2004) Spectrophotometric determination of Rauwolfia alkaloids: estimation of reserpine in pharmaceuticals. Anal Sci 20:571–573. doi:10.2116/analsci.20.571
Srivastava GP, Li P, Liu J, Xu D (2010) Identification of transcription factor’s targets using tissue-specific transcriptomic data in Arabidopsis thaliana. BMC Syst Biol. doi:10.1186/1752-0509-4-S2-S2
Sundar AS, Varghese SM, Shameer K et al (2008) STIF: Identification of stress-upregulated transcription factor binding sites in Arabidopsis thaliana. Bioinformation 2:431–437
Suttipanta N, Pattanaik S, Kulshrestha M et al (2011) The transcription factor CrWRKY1 positively regulates the terpenoid indole alkaloid biosynthesis in Catharanthus roseus. Plant Physiol 157:2081–2093. doi:10.1104/pp.111.181834
The UniProt Consortium (2007) The Universal Protein Resource (UniProt). Nucleic Acids Res 35:D193–D197. doi:10.1093/nar/gkl929
Vakil RJ (1955) Rauwolfia serpentina in the treatment of high blood pressure: a review of the literature. Circulation 12:220–229. doi:10.1161/01.CIR.12.2.220
Van der Fits L, Memelink J (2000) ORCA3, a jasmonate-responsive transcriptional regulator of plant primary and secondary metabolism. Science (80- ) 289:295–297. doi: 10.1126/science.289.5477.295
Van Dijk ADJ, Morabito G, Fiers M et al (2010) Sequence motifs in MADS transcription factors responsible for specificity and diversification of protein-protein interaction. PLoS Comput Biol 6:e1001017. doi:10.1371/journal.pcbi.1001017
Van Dongen S (2008) Graph clustering via a discrete uncoupling process. SIAM J Matrix Anal Appl 30:121–141. doi:10.1137/040608635
Vom Endt D, Kijne JW, Memelink J (2002) Transcription factors controlling plant secondary metabolism: what regulates the regulators. Phytochemistry 61:107–114. doi:10.1016/S0031-9422(02)00185-1
Wang D, Zhang C, Hearn DJ et al (2010) Identification of transcription-factor genes expressed in the Arabidopsis female gametophyte. BMC Plant Biol 10:110. doi:10.1186/1471-2229-10-110
Wei J, van Loon JJA, Gols R et al (2014) Reciprocal crosstalk between jasmonate and salicylate defence-signalling pathways modulates plant volatile emission and herbivore host-selection behaviour. J Exp Bot 65:3289–3298. doi:10.1093/jxb/eru181
Weng J-K, Noel JP (2012) The remarkable pliability and promiscuity of specialized metabolism. Cold Spring Harb Symp Quant Biol 77:309–320. doi:10.1101/sqb.2012.77.014787
Wink M (2010) Annual plant reviews, functions and biotechnology of plant secondary metabolites, 2nd edn. 424
Yang C-Q, Fang X, Wu X-M et al (2012a) Transcriptional regulation of plant secondary metabolism. J Integr Plant Biol 54:703–12. doi:10.1111/j.1744-7909.2012.01161.x
Yang J, Osman K, Iqbal M et al (2012b) Inferring the Brassica rapa interactome using protein-protein interaction data from Arabidopsis thaliana. Front Plant Sci 3:297. doi:10.3389/fpls.2012.00297
Yang LT, Qi YP, Bin LY et al (2013) iTRAQ protein profile analysis of Citrus sinensis roots in response to long-term boron-deficiency. J Proteomics 93:179–206. doi:10.1016/j.jprot.2013.04.025
Yonekura-Sakakibara K, Tohge T, Matsuda F et al (2008) Comprehensive flavonol profiling and transcriptome coexpression analysis leading to decoding gene-metabolite correlations in Arabidopsis. Plant Cell 20:2160–2176. doi:10.1105/tpc.108.058040
Zarate SI, Kempema LA, Walling LL (2007) Silverleaf whitefly induces salicylic acid defenses and suppresses effectual jasmonic acid defenses. Plant Physiol 143:866–875. doi: 10.1104/pp.106.090035
Zhang L, Yu S, Luo KZ et al (2012) Identification of gene modules associated with drought response in rice by network-based analysis. PLoS One 7:e33748. doi:10.1371/journal.pone.0033748
Zhu J-Y, Sun Y, Wang Z-Y (2012) Genome-wide identification of transcription factor-binding sites in plants using chromatin immunoprecipitation followed by microarray (ChIP-chip) or sequencing (ChIP-seq). Methods Mol Biol 876:173–188. doi:10.1007/978-1-61779-809-2_14
Ziegler J, Facchini PJ (2008) Alkaloid Biosynthesis: metabolism and trafficking. Annu Rev Plant Biol 59:735–769. doi:10.1146/annurev.arplant.59.032607.092730
Acknowledgments
We acknowledge the computational infrastructure provided in the form of project MLP0076 by CSIR-Institute of Himalayan Bioresource Technology (CSIR-IHBT), a constituent national laboratory of Council of Scientific and Industrial Research, India, and Department of Biotechnology, Government of India for infrastructural support in the form of Bioinformatics Infrastructure Facility (BIF) as well. The authors are thankful to Dr. Paramvir Singh Ahuja for encouragement and constant support. Shivalika Pathania is grateful to the Department of Science and Technology (DST) for INSPIRE fellowship. We are also thankful to Ashwani Jha and Vinay Randhawa for technical help in manuscript preparation. The CSIR-IHBT communication number for this article is 3777.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Table S1
(XLSX 20 kb)
Table S2
(XLSX 10090 kb)
Table S3
(DOCX 668 kb)
Table S4
(DOCX 15 kb)
Table S5
(XLSX 53 kb)
Table S6
(XLSX 43 kb)
Table S7
(XLSX 36 kb)
Table S8
(XLSX 36 kb)
Table S9
(DOCX 13 kb)
Fig. S1
Gene ontology (GO) annotation of complete R. serpentina transcriptome. Pie chart representing GO-based annotation of complete transcriptome for a biological process and b molecular function category (GIF 92 kb)
Fig. S2
Gene ontology (GO) and KEGG pathways annotation of complete R. serpentina transcriptome. Pie chart representing a GO-based annotation for cellular component and b KEGG pathways annotation of complete transcriptome (GIF 88 kb)
Fig. S3
Threshold selection. a The actual number of edges and all possible edges among non-singleton nodes as a function of PCC cutoff values. b The actual number of edges and all nodes among the non-singleton nodes as a function of PCC cutoff values. The “igraph” library of R package is used to obtain these plots (GIF 13 kb)
Fig. S4
Weighted co-expression network follows power law degree distribution. A data set from the weighted co-expression network is represented with black filled circles, and the degree distribution adheres to a power law as all these circles lie on or close to the red line which is the graph of a function of the form ax-k. The “igraph” library of R package is used to obtain this plot (GIF 12 kb)
Fig. S5
Hierarchical tree representing significantly enriched GO terms for ABI3VP1 TF family. These over-represented GO terms for biological process category are obtained using agriGO. Each GO term represented by box are labeled by their GO ID, term definition, and statistical information. Degree of color saturation of a box is positively correlated to the enrichment level of the term (GIF 48 kb)
Fig. S6
Hierarchical tree representing significantly enriched GO terms for bHLH TF family. These over-represented GO terms for biological process category are obtained using agriGO. Each GO term represented by box are labeled by their GO ID, term definition, and statistical information. Degree of color saturation of a box is positively correlated to the enrichment level of the term (GIF 43 kb)
Fig. S7
Hierarchical tree representing significantly enriched GO terms for HB TF family. These over-represented GO terms for biological process category are obtained using agriGO. Each GO term represented by box are labeled by their GO ID, term definition, and statistical information. Degree of color saturation of a box is positively correlated to the enrichment level of the term (GIF 30 kb)
Fig. S8
Hierarchical tree representing significantly enriched GO terms for MYB TF family. These over-represented GO terms for biological process category are obtained using agriGO. Each GO term represented by box are labeled by their GO ID, term definition, and statistical information. Degree of color saturation of a box is positively correlated to the enrichment level of the term (GIF 58 kb)
Fig. S9
Hierarchical tree representing significantly enriched GO terms for MYB-related TF family. These over-represented GO terms for biological process category are obtained using agriGO. Each GO term represented by box are labeled by their GO ID, term definition, and statistical information. Degree of color saturation of a box is positively correlated to the enrichment level of the term (GIF 55 kb)
Fig. S10
Hierarchical tree representing significantly enriched GO terms for WRKY TF family. These over-represented GO terms for biological process category are obtained using agriGO. Each GO term represented by box are labeled by their GO ID, term definition, and statistical information. Degree of color saturation of a box is positively correlated to the enrichment level of the term (GIF 51 kb)
Fig. S11
Hierarchical tree representing significantly enriched GO terms for -EREBP TF family. These over-represented GO terms for biological process category are obtained using agriGO. Each GO term represented by box are labeled by their GO ID, term definition, and statistical information. Degree of color saturation of a box is positively correlated to the enrichment level of the term (GIF 33 kb)
Fig. S12
Hierarchical tree representing significantly enriched GO terms in enrichment analysis of bZIP TF family. These over-represented GO terms for biological process category are obtained using agriGO. Each GO term represented by box are labeled by their GO ID, term definition, and statistical information. Degree of color saturation of a box is positively correlated to the enrichment level of the term (GIF 7 kb)
Fig. S13
Hierarchical graph representing significantly enriched GO terms for MADS TF family. These over-represented GO terms for biological process category are obtained using agriGO. Each GO term represented by box are labeled by their GO ID, term definition, and statistical information. Degree of color saturation of a box is positively correlated to the enrichment level of the term (GIF 6 kb)
Fig. S14
Clustering of weighted co-expression network. A total of 42 modules are obtained from clustering of weighted co-expression network using MCL algorithm. Colored filled circles representing TFs in each module. Cytoscape is used for visualization of the network (GIF 89 kb)
Fig. S15
Heat map of transcripts in four significant modules (1–4) using expression data of different tissues. Heat map is representing tissue-specific expression of transcripts in significant modules: a module 1 in roots, b module 2 in young leaves, c module 3 in flower, and d module 4 in mature leaves, where average expression is calculated based on normalized transcriptomics data. The “gplots” library of R package is used to plot heat map (GIF 100 kb)
Fig. S16
Top most enriched KEGG pathway annotation for four significant modules (1–4). Each pie segment is labeled with significant KEGG pathway, and the percentage fraction of annotations associated with that particular pathway. a Module 1, b module 2, c module 3, and d module 4 (GIF 65 kb)
Fig. S17
Hierarchical graph representing significantly enriched GO terms for module 2. These over-represented GO terms for biological process category are obtained using agriGO. Each GO term represented by box are labeled by their GO ID, term definition, and statistical information. Degree of color saturation of a box is positively correlated to the enrichment level of the term (GIF 20 kb)
Fig. S18
Hierarchical tree representing significantly enriched GO terms for module 3. These over-represented GO terms for biological process category are generated through agriGO. Each GO term represented by box are labeled by their GO ID, term definition, and statistical information. Degree of color saturation of a box is positively correlated to the enrichment level of the term (GIF 40 kb)
Fig. S19
Hierarchical tree representing significantly enriched GO terms for module 4. These over-represented GO terms for biological process category are obtained using agriGO. Each GO term represented by box are labeled by their GO ID, term definition, and statistical information. Degree of color saturation of a box is positively correlated to the enrichment level of the term (GIF 28 kb)
Fig. S20
Network representing the significantly enriched pathways in module 1. GO terms-based network is obtained from enrichment analysis against reference model A. thaliana for module 1. Functionally grouped pathways are mainly found to be associated to monocarboxylic acid biosynthetic process, hormone-mediated signaling pathway, and defense response. ClueGO plugin is used to generate this GO terms-based network (GIF 100 kb)
Fig. S21
Network representing the significantly enriched pathways in module 2. GO terms-based network is obtained from enrichment analysis against reference model Arabidopsis thaliana. Functionally grouped pathways are mainly found to be associated to “regulation of gene expression, epigenetic,” “RNA metabolic process,” and “macromolecular modification” which also complements agriGO enrichment result. ClueGO, plugin in cytoscape, is used to generate this GO terms-based network. (GIF 95 kb)
Fig. S22
Network representing the significantly enriched pathways in module 3. GO terms-based network is obtained from enrichment analysis against reference model Arabidopsis thaliana. Functionally grouped pathways are mainly found to be associated to “pollen exine formation,” “stamen development,” and “external encapsulating structure organization” which also complements agriGO enrichment result. ClueGO, plugin in cytoscape, is used to generate this GO terms-based network (GIF 27 kb)
Fig. S23
Network representing the significantly enriched pathways in module 4. GO terms-based network is obtained from enrichment analysis against reference model Arabidopsis thaliana. Functionally grouped pathways are mainly found to be associated to “photosynthesis”, “plastid organization,” and “monocarboxylic acid biosynthetic process” which also complements agriGO enrichment result. ClueGO, plugin in cytoscape, is used to generate this GO terms-based network (GIF 80 kb)
Rights and permissions
About this article
Cite this article
Pathania, S., Acharya, V. Computational Analysis of “-omics” Data to Identify Transcription Factors Regulating Secondary Metabolism in Rauvolfia serpentina . Plant Mol Biol Rep 34, 283–302 (2016). https://doi.org/10.1007/s11105-015-0919-1
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11105-015-0919-1