Abstract
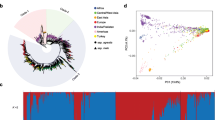
Melon is one of the most important fruit crops in China, and has been the focus of much effort in breeding new cultivars but only limited effort in basic studies (e.g., germplasm collection, assessment, and utilization). In this study, we assessed the genetic diversity and population structure in a large melon collection (570 Chinese accessions and 42 exotic accessions) using 26 microsatellite markers distributed evenly in the genome. A total of 355 alleles, a high level of gene diversity (0.713), and a low observed heterozygosity (0.177) were detected among the whole panel. Using a model-based population structure analysis, all accessions were assigned to one of two main groups (P1 and P2), which were largely in line with their subspecies classification and geographic distribution. P1 comprised 279 accessions, most of which were from Northwest China, while P2 comprised 333 accessions, most of which were from the northeast, central, and east regions in China. Each of the two main groups was further subdivided into two subgroups that had different fruit features. Principal component analysis also gave similar results in positioning the 612 melon accessions. AMOVA, pairwise FST, and Nei’s genetic distance confirmed the differentiation between the groups and subgroups. The accessions from Northwest China revealed a high level of genetic diversity, as did accessions from Northeast, Central, and East China. Subsequently, we constructed a core collection representing 19.4 % of the whole panel and showing 100 % coverage of alleles. Our results provide an effective aid for future germplasm conservation and association genetics as well as development of appropriate breeding strategies in melon.




Similar content being viewed by others
References
Aierken Y, Akashi Y, Phan TPN, Halidan Y, Tanaka K, Long B, Nishida H, Long CL, Wu MZ, Kato K (2011) Molecular analysis of the genetic diversity of Chinese Hami melon and its relationship to the melon germplasm from central and south Asia. J Jpn Soc Hortic Sci 80:52–65
Akashi Y, Fukuda N, Wako T, Masuda M, Kato K (2002) Genetic variation and phylogenetic relationships in East and South Asian melons, Cucumis melo L., based on the analysis of five isozymes. Euphytica 125:385–396
Brown AHD (1989) Core collections: a practical approach to genetic resources management. Genome 31:818–824
Creste S, Neto AT, Figueira A (2001) Detection of single sequence repeat polymorphisms in denaturing polyacrylamide sequencing gels by silver staining. Plant Mol Biol Rep 19:229–306
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software structure: a simulation study. Mol Ecol 14:2611–2620
Excoffier L, Laval G, Schneider S (2005) Arlequin ver. 3.0: an integrated software package for population genetics data analysis. Evol Bioinform 1:47–50
Gao P, Ma H, Luan F, Song H (2012) DNA fingerprinting of Chinese melon provides evidentiary support of seed quality appraisal. Plos One 7(12):e52431
Gonzalo MJ, Oliver M, Garcia-Mas J, Monfort A, Dolcet-Sanjuan R, Katzir N, Arús P, Monforte AJ (2005) Simple-sequence repeat markers used in merging linkage maps of melon (Cucumis melo L.). Theor Appl Genet 110:802–811
Gupta PK, Rustgi S, Kulwal PL (2005) Linkage disequilibrium and association studies in higher plants: present status and future prospects. Plant Mol Biol 57:461–485
Fukino N, Ohara Y, Monforte AJ, Sugiyama M, Sakata Y, Kunihisa M, Matsumoto S (2008) Identification of QTLs for resistance to powdery mildew and SSR markers diagnostic for powdery mildew resistance genes in melon (Cucumis melo L.). Theor Appl Genet 118:165–175
Ingvarsson PK, Street NR (2011) Association genetics of complex traits in plants. New Phytol 189:909–922
Innark P, Khanobdee C, Samipak S, Jantasuriyarat C (2013) Evaluation of genetic diversity in cucumber (Cucumis sativus L.) germplasm using agro-economic traits and microsatellite markers. Sci Hortic 162:278–284
Kaçar YA, Simsek O, Solmaz I, Sari N, Mendi YY (2012) Genetic diversity among melon accessions (Cucumis melo) from Turkey based on SSR markers. Genet Mol Res 11:622–4631
Kim KW, Chung HK, Cho GT, Ma KH, Chandrabalan D, Gwag JG, Kim TS, Cho EG, Park YJ (2007) Power Core: A program applying the advanced M strategy with a heuristic search for establishing core sets. Bioinformatics 23:2155–2162
Kirkbride JH (1993) Biosystematics monograph of the genus Cucumis (Cucurbitaceae). Parkway, Boone, NC
Hu JB, Ma SW, Jian ZH, Wang JM, Li Q, Su Y (2013) Analysis of genetic diversity of Chinese melon (Cucumis melo L.) germplasm resource based on morphological characters. J Plant Genet Resour 14:612–619
Kong Q, Xiang C, Yang J, Yu Z (2011) Genetic variations of Chinese melon landraces investigated with EST-SSR markers. Hortic Environ Biotechnol 52:163–169
Li H, Younas M, Wang X, Li X, Chen L, Zhao B, Chen X, Xu J, Hou F, Hong B, Liu G, Zhao H, Wu X, Du H, Wu J, Liu K (2013) Development of a core set of single-locus SSR markers for allotetraploid rapeseed (Brassica napus L.). Theor Appl Genet 126:937–47
Lindholm E, Hodge SE, Greenberg DA (2004) Comparative informativeness for linkage of multiple SNPs and single microsatellites. Hum Hered 58:164–170
Liu K, Muse SV (2005) PowerMarker: an integrated analysis environment for genetic marker analysis. Bioinformatics 21:2128–2129
Luan F, Delannay I, Staub JE (2008) Chinese melon (Cucumis melo L.) diversity analyses provide strategies for germplasm curation, genetic improvement, and evidentiary support of domestication patterns. Euphytica 164:445–461
McCreight JD, Staub JE, López-Sesé A, Chung SM (2004) Isozyme variation in Indian and Chinese melon (Cucumis melo L.) germplasm collections. J Am Soc Hortic Sci 129:811–818
Monforte AJ, Garcia-Mas J, Arús P (2003) Genetic variability in melon based on microsatellite variation. Plant Breed 122:153–157
Murray MG, Thompson WF (1980) Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res 8:4321–4325
Nakata E, Staub JE, López-Sesé AI, Katzir N (2005) Genetic diversity of Japanese melon cultivars (Cucumis melo L.) as assessed by random amplified polymorphic DNA and simple sequence repeat markers. Genet Resour Crop Evol 52:405–419
Nantoumé AD, Andersen SB, Jensen BD (2013) Genetic differentiation of watermelon landrace types in Mali revealed by microsatellite (SSR) markers. Genet Resour Crop Evol 60:2129–2141
Nei M (1978) Estimation of average heterozygosity and genetic distance from a small number of individuals. Genetics 89:583–590
Odong TL, Jansen J, van Eeuwijk FA, van Hintum TJL (2013) Quality of core collections for effective utilisation of genetic resources review, discussion and interpretation. Theor Appl Genet 126:289–305
Pitrat M (2008) Melon. In: Prohens J, Nuez F (eds) Handbook of plant breeding, vol 1. Springer, New York, pp 283–315
Pitrat M, Chauvet M, Foury C (1999) Diversity, history and production of cultivated cucurbits. Acta Hortic (ISHS) 492:21–28
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure from multilocus genotype data. Genetics 155:945–959
Raghami M, López-Sesé AI, Hasandokht MR, Zamani Z, Moghadam MRF, Kashi A (2014) Genetic diversity among melon accessions from Iran and their relationships with melon germplasm of diverse origins using microsatellite markers. Plant Syst Evol 300:139–151
Rao ES, Kadirvel P, Symonds RC, Geethnajali S, Ebert AW (2011) Using SSR markers to map genetic diversity and population structure of Solanum pimpinellifolium for development of a core collection. Plant Genet Resour 10:38–48
Remington DL, Thornsberry JM, Matsuoka Y, Wilson LM, Whitt SR, Doebley J, Kresovich S, Goodman MM, Buckler ES (2001) Structure of linkage disequilibrium and phenotypic associations in the maize genome. Proc Natl Acad Sci USA 98:11479–11484
Renner SS, Schaefer H, Kocyan A (2007) Phylogenetics of Cucumis (Cucurbitaceae): Cucumber (C. sativus) belongs in an Asian/Australian clade far from melon (C. melo). BMC Evol Biol 7:58
Robinson RW, Decker-Walters D (1997) Cucurbits. Cab International, New York
Roy A, Bal SS, Fergany M, Kaur S, Singh H, Malik AA, Singh J, Monforte AJ, Dhillon NPS (2012) Wild melon diversity in India (Punjab State). Genet Resour Crop Evol 59:755–767
Sebastian P, Schaefer H, Telford IRH, Renner SS (2010) Cucumber (Cucumis sativus) and melon (C. melo) have numerous wild relatives in Asia and Australia, and the sister species of melon is from Australia. Proc Natl Acad Sci USA 107:14269–14273
Shen Y, Luan F, Chen K, Cui X, Staub JE (2007) Genetic diversity of Chinese thin-Skinned melon cultivars (Cucumis melo L.) based on simple sequence repeat markers. Acta Hortic (ISHS) 763:169–176
Staub JE, Luan F, Delannay I (2007) Genetic diversity in Chinese melon (Cucumis melo L.). Hortic Sci 42:856–857
Tomason Y, Nimmakayala P, Levi A, Reddy UK (2013) Map-based molecular diversity, linkage disequilibrium and association mapping of fruit traits in melon. Mol Breed 31:829–841
Van Inghelandt D, Melchinger AE, Lebreton C, Stich B (2010) Population structure and genetic diversity in a commercial maize breeding program assessed with SSR and SNP markers. Theor Appl Genet 120:1289–1299
Xanthopoulou A, Ganopoulos I, Tsaballa A, Nianiou-Obeidat I, Kalivas A, Tsaftaris A, Madesis P (2014) Summer squash identification by High-Resolution-Melting (HRM) analysis using gene-based EST-SSR molecular markers. Plant Mol Biol Rep 32:395–405
Walters TW (1989) Historical overview of domesticated plants in China with special emphasis on the Cucurbitaceae. Econ Bot 43:297–313
Wang JY, Mou YK (1980) The excavations of the Qianshanyang site in Wuxing country, Zhejiang (in Chinese). Archaeology 4:353–358
Yeh FC, Boyle TJB (1997) Population genetic analysis of co-dominant and dominant markers and quantitative traits. Belg J Bot 129:157
Zhang N, Luan F, Gao P (2012) The study of disease resistant, morphlogical characteristics and genetic relationship using SSR markers in four materials of wild cucurbit (in Chinese with an English abstract). Acta Hortic Sin 39:905–914
Acknowledgments
We are grateful to Prof. Z.-F. Yan, College of Horticulture of Henan Agricultural University, for helpful discussions and critical reading of this manuscript. We also thank Zhengzhou Fruit Research Institute of Chinese Academy of Agriculture Sciences for providing melon germplasm resources. The research was supported by the National Natural Science Foundation of China (Grant No. 31101544).
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Table S1
List of the 612 melon accessions used in the present study. The groups (P1 and P2) and subgroups (G1, G2, G3, and G4) for these accessions were noted as well as the accession types. Accession type: B breeding line, C commercial hybrid or experimental hybrid, L landrace, W wild accession. Of these accessions, those chosen as the core collection were recorded as “Y” (XLS 118 kb)
Table S2
Description of the horticultural traits of 22 diverse melon accessions used for screening of SSR markers (XLS 20 kb)
Table S3
Characteristics of the 26 SSR markers in the present study (XLS 19 kb)
Fig. S1
Determination of LnP (D) and ΔK in the total panel and inferred groups. (a) the total panel; (b) the P1 group; (c) the P2 group. The blue and red curves represent LnP (D) and ΔK, respectively. The bar indicates standard deviation (GIF 77 kb)
Rights and permissions
About this article
Cite this article
Hu, J., Wang, P., Su, Y. et al. Microsatellite Diversity, Population Structure, and Core Collection Formation in Melon Germplasm. Plant Mol Biol Rep 33, 439–447 (2015). https://doi.org/10.1007/s11105-014-0757-6
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11105-014-0757-6