Abstract
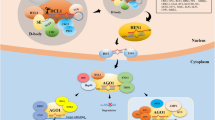
MicroRNAs (miRNAs) are small ∼20–24 nt species of non-coding RNAs that modulate plant gene expression by means of gene silencing through sequence-specific inhibition of target mRNAs. MiRNAs derive from pol-II transcription of non-coding genes that are precisely processed in nuclear Dicing bodies by a microprocessor complex (dicer-like1–serrate–hyponastic leaves 1: DCL1-SE-HYL1), which recognizes stem-loop secondary-structure features of primary precursor miRNA transcripts (pri-miRNA). The proper processing of the pri-miRNAs results in a double-stranded small RNA that will eventually exit the nucleus and be loaded predominantly onto the effector complex Argonaute1 (Ago1). The single-stranded mature miRNA will guide AGO1, leading to cleavage or translational arrest of complementary mRNAs. MiRNA steady-state levels and activity are regulated not only by transcription rate of precursor transcripts, but also by direct degradation mediated by small RNA degrading nuclease1 (SDN1). miRNAs are retailored by 3′ editing through 2-O-methylation, uridylation and adenlylation, involving Hua enhancer1 (HEN1), HEN1 suppressor1 (HESO1) and probably the exosome—a phenomenon that has been elucidated only scarcely to date in Arabidopsis. MiRNA activity is involved not only in plant development, but also in signaling, abiotic stresses such as drought, heat and metal toxicity, pathogen interaction and symbiotic relationship regulation, among others. The engineering of miRNAs is paving the way to next-generation plant biotechnology by means of over-expression of natural miRNAs, generation of artificial microRNAs and inhibition of miRNA activity by target mimicry. This review highlights the importance of miRNAs in plant sciences by describing the latest updates in this research field.



Similar content being viewed by others
References
Allen E, Xie Z, Gustafson AM, Carrington JC (2005) microRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell 121:207–221
Auer C (2011) Small RNAs for crop improvement: applications and considerations for ecological risk assessments. In: Erdmann VA, Barciszewski J (eds) Non-coding RNAs in plants. Springer, Berlin, pp 461–484
Auer C, Frederick R (2009) Crop improvement using small RNAs: applications and predictive ecological risk assessments. Trends Biotechnol 27:644–651
Axtell MJ, Bowman JL (2008) Evolution of plant microRNAs and their targets. Trends Plant Sci 13:343–349
Axtell MJ, Jan C, Rajagopalan R, Bartel DP (2006) A two-hit trigger for siRNA biogenesis in plants. Cell 127:565–577
Balmer D, Mauch-Mani B (2013) Small yet mighty microRNAs in plant-microbe interactions. MicroRNA 2(1):73–80
Bazin J, Khan GA, Combier J-P et al (2013) miR396 affects mycorrhization and root meristem activity in the legume Medicago truncatula. Plant J 74:920–934
Bazzini AA, Hopp HE, Beachy RN, Asurmendi S (2007) Infection and coaccumulation of tobacco mosaic virus proteins alter microRNA levels, correlating with symptom and plant development. Proc Natl Acad Sci USA 104:12157–12162
Bergonzi S, Albani MC, Loren V, van Themaat E et al (2013) Mechanisms of age-dependent response to winter temperature in perennial flowering of Arabis alpina. Science 340:1094–1097
Bologna NG, Schapire AL, Zhai J et al (2013) Multiple RNA recognition patterns during microRNA biogenesis in plants. Genome Res 23:1675–1689
Brodersen P, Sakvarelidze-Achard L, Bruun- M (2008) Widespread translational inhibition by plant miRNAs and siRNAs. Science 320:1185–1190
Brousse C, Liu Q, Beauclair L, et al. (2014) A non-canonical plant microRNA target site. Nucleic Acids Res. doi: 10.1093/nar/gku157
Burko Y, Shleizer-Burko S, Yanai O et al (2013) A role for APETALA1/fruitfull transcription factors in tomato leaf development. Plant Cell 25:2070–2083
Campo S, Peris-Peris C, Siré C et al (2013) Identification of a novel microRNA (miRNA) from rice that targets an alternatively spliced transcript of the Nramp6 (Natural resistance-associated macrophage protein 6) gene involved in pathogen resistance. New Phytol 199:212–227
Carbonell A, Fahlgren N, Garcia-Ruiz H et al (2012) Functional analysis of three Arabidopsis ARGONAUTES using slicer-defective mutants. Plant Cell 24:3613–3629
Cesana M, Daley GQ (2013) Deciphering the rules of ceRNA networks. Proc Natl Acad Sci USA 110:7112–7113
Chaabane S, Liu R, Chinnusamy V et al (2013) STA1, an Arabidopsis pre-mRNA processing factor 6 homolog, is a new player involved in miRNA biogenesis. Nucleic Acids Res 41:1984–1997
Chapman EJ, Prokhnevsky AI, Gopinath K et al (2004) Viral RNA silencing suppressors inhibit the microRNA pathway at an intermediate step. Genes Dev 18:1179–1186
Chen X (2012) Small RNAs in development—insights from plants. Curr Opin Genet Dev 22:361–7
Chen H-M, Chen L-T, Patel K et al (2010) 22-nucleotide RNAs trigger secondary siRNA biogenesis in plants. Proc Natl Acad Sci USA 107:15269–15274
Chen L, Zhang Y, Ren Y et al (2012) Genome-wide identification of cold-responsive and new microRNAs in Populus tomentosa by high-throughput sequencing. Biochem Biophys Res Commun 417:892–896
Chen X, Zen K, Zhang C-Y (2013) Reply to Lack of detectable oral bioavailability of plant microRNAs after feeding in mice. Nat Biotechnol 31:967–969
Cho SH, Coruh C, Axtell MJ (2012) miR156 and miR390 regulate tasiRNA accumulation and developmental timing in Physcomitrella patens. Plant Cell 24:4837–4849
Cloonan N, Wani S, Xu Q et al (2011) MicroRNAs and their isomiRs function cooperatively to target common biological pathways. Genome Biol 12:R126
Cuperus J, Carbonell A, Fahlgren NH (2010) Unique functionality of 22-nt miRNAs in triggering RDR6-dependent siRNA biogenesis from target transcripts in Arabidopsis. Nat Struct Mol Biol 17:997–1003
De Felippes FF, Wang J, Weigel D (2012) MIGS: miRNA-induced gene silencing. Plant J 70:541–547
De Oliveira LFV, Christoff AP, Margis R (2013) isomiRID: a framework to identify microRNA isoforms. Bioinformatics 29:2521–2523
Debernardi JM, Rodriguez RE, Mecchia MA, Palatnik JF (2012) Functional specialization of the plant miR396 regulatory network through distinct microRNA-target interactions. PLoS Genet 8:e1002419
Dickinson B, Zhang Y, Petrick JS et al (2013) Lack of detectable oral bioavailability of plant microRNAs after feeding in mice. Nat Biotechnol 31:965–967
Ding J, Li D, Ohler U et al (2012) Genome-wide search for miRNA-target interactions in Arabidopsis thaliana with an integrated approach. BMC Genomics 13(Suppl 3):S3
Ding Y, Tao Y, Zhu C (2013) Emerging roles of microRNAs in the mediation of drought stress response in plants. J Exp Bot 64:3077–3086
Fischer JJ, Beatty PH, Good AG, Muench DG (2013) Manipulation of microRNA expression to improve nitrogen use efficiency. Plant Sci 210:70–81
Franco-zorrilla M, Puga I, Todesco M et al (2007) Target mimicry provides a new mechanism for regulation of microRNA activity. Nat Genet 39:1033–1037
Fu C, Sunkar R, Zhou C et al (2012) Overexpression of miR156 in switchgrass (Panicum virgatum L.) results in various morphological alterations and leads to improved biomass production. Plant Biotechnol J 10:443–452
Giacomelli JI, Weigel D, Chan RL, Manavella PA (2012) Role of recently evolved miRNA regulation of sunflower HaWRKY6 in response to temperature damage. New Phytol 195:766–773
Guan Q, Lu X, Zeng H et al (2013) Heat stress induction of miR398 triggers a regulatory loop that is critical for thermotolerance in Arabidopsis. Plant J 74:840–851
Gupta OP, Sharma P, Gupta RK, Sharma I (2014) MicroRNA mediated regulation of metal toxicity in plants: present status and future perspectives. Plant Mol Biol 84:1–18
Hajheidari M, Koncz C, Eick D (2013) Emerging roles for RNA polymerase II CTD in Arabidopsis. Trends Plant Sci 18:633–643
Hauser F, Chen W, Deinlein U et al (2013) A genomic-scale artificial microRNA library as a tool to investigate the functionally redundant gene space in Arabidopsis. Plant Cell 25:2848–2863
Ibrahim F, Rymarquis LA, Kim E-J et al (2010) Uridylation of mature miRNAs and siRNAs by the MUT68 nucleotidyltransferase promotes their degradation in Chlamydomonas. Proc Natl Acad Sci USA 107:3906–3911
Iwata Y, Takahashi M, Fedoroff NV, Hamdan SM (2013) Dissecting the interactions of SERRATE with RNA and DICER-LIKE 1 in Arabidopsis microRNA precursor processing. Nucleic Acids Res 41:9129–9140
Jeong D-H, Park S, Zhai J et al (2011) Massive analysis of rice small RNAs: mechanistic implications of regulated microRNAs and variants for differential target RNA cleavage. Plant Cell 23:4185–4207
Jeong D-H, Thatcher SR, Brown RSH et al (2013) Comprehensive investigation of microRNAs enhanced by analysis of sequence variants, expression patterns, ARGONAUTE loading, and target cleavage. Plant Physiol 162:1225–1245
Jia F, Rock CD (2013) MIR846 and MIR842 comprise a cistronic MIRNA pair that is regulated by abscisic acid by alternative splicing in roots of Arabidopsis. Plant Mol Biol 81:447–460
Jin D, Wang Y, Zhao Y, Chen M (2013) MicroRNAs and their cross-talks in plant development. J Genet Genomics 40:161–70
Jones-Rhoades MW, Bartel DP (2004) Computational identification of plant microRNAs and their targets, including a stress-induced miRNA. Mol Cell 14:787–799
Jones-Rhoades MW, Bartel DP, Bartel B (2006) MicroRNAs and their regulatory roles in plants. Annu Rev Plant Biol 57:19–53
Källman T, Chen J, Gyllenstrand N, Lagercrantz U (2013) A significant fraction of 21-nucleotide small RNA originates from phased degradation of resistance genes in several perennial species. Plant Physiol 162:741–754
Kettles GJ, Drurey C, Schoonbeek H et al (2013) Resistance of Arabidopsis thaliana to the green peach aphid, Myzus persicae, involves camalexin and is regulated by microRNAs. New Phytol 198:1178–1190
Knauer S, Holt AL, Rubio-Somoza I et al (2013) A protodermal miR394 signal defines a region of stem cell competence in the Arabidopsis shoot meristem. Dev Cell 24:125–132
Lafforgue G, Martínez F, Niu Q-W et al (2013) Improving the effectiveness of artificial microRNA (amiR)-mediated resistance against Turnip mosaic virus by combining two amiRs or by targeting highly conserved viral genomic regions. J Virol 87:8254–8256
Li F, Pignatta D, Bendix C et al (2012a) MicroRNA regulation of plant innate immune receptors. Proc Natl Acad Sci USA 109:1790–1795
Li S, Yang X, Wu F, He Y (2012b) HYL1 controls the miR156-mediated juvenile phase of vegetative growth. J Exp Bot 63:2787–98
Li J-F, Chung HS, Niu Y et al (2013a) Comprehensive protein-based artificial microRNA screens for effective gene silencing in plants. Plant Cell 25:1507–1522
Li S, Liu L, Zhuang X et al (2013b) MicroRNAs inhibit the translation of target mRNAs on the endoplasmic reticulum in Arabidopsis. Cell 153:562–574
Li X, Bian H, Song D et al (2013c) Flowering time control in ornamental gloxinia (Sinningia speciosa) by manipulation of miR159 expression. Ann Bot 111:791–799
Liang G, He H, Yu D (2012) Identification of nitrogen starvation-responsive microRNAs in Arabidopsis thaliana. PLoS One 7:e48951
Liu C, Axtell MJ, Fedoroff NV (2012) The helicase and RNaseIIIa domains of Arabidopsis Dicer-Like1 modulate catalytic parameters during microRNA biogenesis. Plant Physiol 159:748–758
Liu Q, Yan Q, Liu Y et al (2013) Complementation of HYPONASTIC LEAVES1 by double-strand RNA-binding domains of DICER-LIKE1 in nuclear dicing bodies. Plant Physiol 163:108–117
Liu Q, Wang F, Axtell MJ (2014) Analysis of complementarity requirements for plant microRNA targeting using a Nicotiana benthamiana quantitative transient assay. Plant Cell. doi: 10.1105/tpc.113.120972
Lu S, Sun Y, Chiang VL (2009) Adenylation of plant miRNAs. Nucleic Acids Res 37:1878–1885
Ma C, Lu Y, Bai S et al (2014) Cloning and characterization of miRNAs and their targets, including a novel miRNA-targeted NBS-LRR protein class gene in apple (Golden Delicious). Mol Plant 7:218–230
Manacorda CA, Mansilla C, Debat HJ et al (2013) Salicylic acid determines differential senescence produced by two Turnip mosaic virus strains involving reactive oxygen species and early transcriptomic changes. Mol Plant Microbe Interact 26:1486–1498
Manavella PA, Hagmann J, Ott F et al (2012a) Fast-forward genetics identifies plant CPL phosphatases as regulators of miRNA processing factor HYL1. Cell 151:859–870
Manavella PA, Koenig D, Weigel D (2012b) Plant secondary siRNA production determined by microRNA-duplex structure. Proc Natl Acad Sci USA 109:2461–2466
Manavella PA, Koenig D, Rubio-Somoza I et al (2013) Tissue-specific silencing of Arabidopsis SU(VAR)3-9 HOMOLOG8 by miR171a. Plant Physiol 161:805–812
Martínez F, Elena SF, Daròs J-A (2013) Fate of artificial microRNA-mediated resistance to plant viruses in mixed infections. Phytopathology 103:870–876
McHale M, Eamens AL, Finnegan EJ, Waterhouse PM (2013) A 22-nt artificial microRNA mediates widespread RNA silencing in Arabidopsis. Plant J 76:519–529
Mi S, Cai T, Hu Y et al (2008) Sorting of small RNAs into Arabidopsis argonaute complexes is directed by the 5′ terminal nucleotide. Cell 133:116–127
Molnár A, Schwach F, Studholme DJ et al (2007) miRNAs control gene expression in the single-cell alga Chlamydomonas reinhardtii. Nature 447:1126–1129
Neilsen CT, Goodall GJ, Bracken CP (2012) IsomiRs–the overlooked repertoire in the dynamic microRNAome. Trends Genet 28:544–549
Niu Q-W, Lin S-S, Reyes JL et al (2006) Expression of artificial microRNAs in transgenic Arabidopsis thaliana confers virus resistance. Nat Biotechnol 24:1420–1428
Palatnik JF, Allen E, Wu X, et al. (2003) Control of leaf morphogenesis by microRNAs. Nature 425:257–263
Palukaitis P, Groen SC, Carr JP (2013) The Rumsfeld paradox: some of the things we know that we don’t know about plant virus infection. Curr Opin Plant Biol 16:513–519
Park W, Li J, Song R et al (2002) CARPEL FACTORY, a dicer homolog, and HEN1, a novel protein, Act in microRNA metabolism in Arabidopsis thaliana. Curr Biol 12:1484–1495
Peragine A, Yoshikawa M, Park MY, Poethig RS (2005) A pathway for the biogenesis of trans-acting siRNAs in Arabidopsis. A pathway for the biogenesis of trans-acting siRNAs in Arabidopsis. Genes Dev 19:2164–2175
Pumplin N, Voinnet O (2013) RNA silencing suppression by plant pathogens: defence, counter-defence and counter-counter-defence. Nat Rev Microbiol 11:745–760
Rajagopalan R, Vaucheret H, Trejo J, Bartel D (2006) A diverse and evolutionarily fluid set of microRNAs in Arabidopsis thaliana. Genes Dev 20:3407–3425
Ramachandran V, Chen X (2008) Degradation of microRNAs by a family of exoribonucleases in Arabidopsis. Science 321:490–1492
Ren G, Chen X, Yu B (2012a) Uridylation of miRNAs by hen1 suppressor1 in Arabidopsis. Curr Biol 22:695–700
Ren G, Xie M, Dou Y et al (2012b) Regulation of miRNA abundance by RNA binding protein TOUGH in Arabidopsis. Proc Natl Acad Sci USA 109:12817–12821
Rhoades MW, Reinhart BJ, Lim LP et al (2002) Prediction of Plant MicroRNA Targets. Cell 110:513–520
Rodrigo G, Elena SF (2013) MicroRNA precursors are not structurally robust but plastic. Genome Biol Evol 5:181–186
Rogers K, Chen X (2012) microRNA biogenesis and turnover in plants. Cold Spring Harb Symp Quant Biol 77:183–194
Rubio-Somoza I, Weigel D (2011) MicroRNA networks and developmental plasticity in plants. Trends Plant Sci 16:258–264
Rubio-Somoza I, Weigel D (2013) Coordination of flower maturation by a regulatory circuit of three microRNAs. PLoS Genet 9:e1003374
Rubio-Somoza I, Weigel D, Franco-Zorilla J-M et al (2011) ceRNAs: miRNA target mimic mimics. Cell 147:1431–1432
Sablok G, Milev I, Minkov G et al (2013) isomiRex: web-based identification of microRNAs, isomiR variations and differential expression using next-generation sequencing datasets. FEBS Lett 587:2629–2634
Salmena L, Poliseno L, Tay Y et al (2011) A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell 146:353–358
Schwab R, Palatnik JF, Riester M et al (2005) Specific effects of microRNAs on the plant transcriptome. Dev Cell 8:517–527
Shivaprasad PV, Chen H-M, Patel K et al (2012) A microRNA superfamily regulates nucleotide binding site-leucine-rich repeats and other mRNAs. Plant Cell 24:859–874
Speth C, Willing E-M, Rausch S et al (2013) RACK1 scaffold proteins influence miRNA abundance in Arabidopsis. Plant J 76:433–445
Tang Z, Zhang L, Xu C et al (2012) Uncovering small RNA-mediated responses to cold stress in a wheat thermosensitive genic male-sterile line by deep sequencing. Plant Physiol 159:721–738
Thieme CJ, Schudoma C, May P, Walther D (2012) Give it AGO: The search for miRNA-Argonaute sorting signals in Arabidopsis thaliana indicates a relevance of sequence positions other than the 5′-position alone. Front Plant Sci 3:272
Todesco M, Rubio-Somoza I, Paz-Ares J, Weigel D (2010) A collection of target mimics for comprehensive analysis of microRNA function in Arabidopsis thaliana. PLoS Genet 6:e1001031
Todesco M, Balasubramanian S, Cao J et al (2012) Natural variation in biogenesis efficiency of individual Arabidopsis thaliana microRNAs. Curr Biol 22:166–170
Turner M, Nizampatnam NR, Baron M et al (2013) Ectopic expression of miR160 results in auxin hypersensitivity, cytokinin hyposensitivity, and inhibition of symbiotic nodule development in soybean. Plant Physiol 162:2042–2055
Varallyay E, Havelda Z (2013) Unrelated viral suppressors of RNA silencing mediate the control of ARGONAUTE1 level. Mol Plant Pathol 14:567–575
Varallyay E, Valoczi A, Agyi A et al (2010) Plant virus-mediated induction of miR168 is associated with repression of ARGONAUTE1 accumulation. EMBO J 29:3507–3519
Vazquez F, Vaucheret H, Rajagopalan R et al (2004) Endogenous trans-acting siRNAs regulate the accumulation of Arabidopsis mRNAs. Mol Cell 16:69–79
Wang J, Czech B, Weigel D (2009) miR156-regulated SPL transcription factors define an endogenous flowering pathway in Arabidopsis thaliana. Cell 138:738–749
Wang L, Song X, Gu L et al (2013) NOT2 proteins promote polymerase II-dependent transcription and interact with multiple MicroRNA biogenesis factors in Arabidopsis. Plant Cell 25:715–727
Wu G, Park MY, Conway SR et al (2009) The sequential action of miR156 and miR172 regulates developmental timing in Arabidopsis. Cell 138:750–759
Wu H-J, Wang Z-M, Wang M, Wang X-J (2013a) Widespread long noncoding RNAs as endogenous target mimics for microRNAs in plants. Plant Physiol 161:1875–1884
Wu X, Shi Y, Li J et al (2013b) A role for the RNA-binding protein MOS2 in microRNA maturation in Arabidopsis. Cell Res 23:645–657
Xia R, Meyers BC, Liu Z et al (2013) MicroRNA superfamilies descended from miR390 and their roles in secondary small interfering RNA Biogenesis in Eudicots. Plant Cell 25:1555–1572
Xie Z, Allen E, Fahlgren N et al (2005) Expression of Arabidopsis MIRNA genes. Plant Physiol 138:2145–2154
Xing S, Salinas M, Garcia-Molina A et al (2013) SPL8 and miR156-targeted SPL genes redundantly regulate Arabidopsis gynoecium differential patterning. Plant J 75:566–577
Xu L, Wang Y, Zhai L et al (2013) Genome-wide identification and characterization of cadmium-responsive microRNAs and their target genes in radish (Raphanus sativus L.) roots. J Exp Bot 64:4271–4287
Yan J, Gu Y, Jia X et al (2012a) Effective small RNA destruction by the expression of a short tandem target mimic in Arabidopsis. Plant Cell 24:415–427
Yan K, Liu P, Wu C-A et al (2012b) Stress-induced alternative splicing provides a mechanism for the regulation of microRNA processing in Arabidopsis thaliana. Mol Cell 48:521–531
Yan Z, Hossain MS, Wang J et al (2013) miR172 regulates soybean nodulation. Mol Plant Microbe Interact 26:1371–1377
Yang L, Xu M, Koo Y et al (2013) Sugar promotes vegetative phase change in Arabidopsis thaliana by repressing the expression of MIR156A and MIR156C. Elife 2:e00260
Yu B, Yang Z, Li J et al (2005) Methylation as a crucial step in plant microRNA biogenesis. Science 307:932–935
Zhai J, Zhao Y, Simon SA et al (2013) Plant microRNAs display differential 3′ truncation and tailing modifications that are ARGONAUTE1 dependent and conserved across species. Plant Cell 25:2417–2428
Zhang H, Li L (2013) SQUAMOSA promoter binding protein-like7 regulated microRNA408 is required for vegetative development in Arabidopsis. Plant J 74:98–109
Zhang L, Hou D, Chen X et al (2011) Exogenous plant MIR168a specifically targets mammalian LDLRAP1: evidence of cross-kingdom regulation by microRNA. Cell Res 22:107–126
Zhang J, Zhang S, Li S et al (2013a) A genome-wide survey of microRNA truncation and 3’ nucleotide addition events in larch (Larix leptolepis). Planta 237:1047–1056
Zhang Y-C, Yu Y, Wang C-Y et al (2013b) Overexpression of microRNA OsmiR397 improves rice yield by increasing grain size and promoting panicle branching. Nat Biotechnol 31:848–852
Zhang X-N, Li X, Liu J-H (2014) Identification of conserved and novel cold-responsive microRNAs in trifoliate orange (Poncirus trifoliata (L.) Raf.) using high-throughput sequencing. Plant Mol Biol Report 32:328–341
Zhao M, Tai H, Sun S et al (2012a) Cloning and characterization of maize miRNAs involved in responses to nitrogen deficiency. PLoS One 7:e29669
Zhao Y, Yu Y, Zhai J et al (2012b) The Arabidopsis nucleotidyl transferase HESO1 uridylates unmethylated small RNAs to trigger their degradation. Curr Biol 22:689–694
Zhao X, Zhang H, Li L (2013) Identification and analysis of the proximal promoters of microRNA genes in Arabidopsis. Genomics 101:187–194
Zhou Z, Wang Z, Li W et al (2013a) Comprehensive analyses of microRNA gene evolution in paleopolyploid soybean genome. Plant J 76:332–344
Zhou C-M, Zhang T-Q, Wang X et al (2013b) Molecular basis of age-dependent vernalization in Cardamine flexuosa. Science 340:1097–1100
Zhu H, Zhou Y, Castillo-González C et al (2013) Bidirectional processing of pri-miRNAs with branched terminal loops by Arabidopsis Dicer-like1. Nat Struct Mol Biol 20:1106–1115
Zou Y, Wang Y, Wang L et al (2013) miR172b controls the transition to autotrophic development inhibited by ABA in Arabidopsis. PLoS One 8:e64770
Zvereva AS, Pooggin MM (2012) Silencing and innate immunity in plant defense against viral and non-viral pathogens. Viruses 4:2578–2597
Acknowledgments
This work was supported by the National Program of Plant Protection, Instituto Nacional de Tecnología Agropecuaria (INTA). The authors would like apologize to colleagues whose work could not be fully cited owing to space constraints.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Debat, H.J., Ducasse, D.A. Plant microRNAs: Recent Advances and Future Challenges. Plant Mol Biol Rep 32, 1257–1269 (2014). https://doi.org/10.1007/s11105-014-0727-z
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11105-014-0727-z