Abstract
Aims
The proliferation of antibiotic resistance genes (ARGs) is a growing public health concern worldwide. Several studies have substantiated the significant role played by the plant microbiome in facilitating the transmission of antibiotic resistance, thereby posing a substantial threat to human health. However, the underlying mechanism of the transmission of antibiotic resistance from soil to plants is still largely unexplored.
Methods
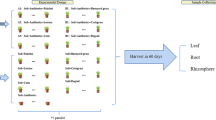
In the current study, we selected Sedum alfredii as a model plant to investigate the occurrence and distribution of ARGs across the soil–plant interface.
Results
Our results showed that 20.34% of ARGs were significantly shifted between rhizosphere and bulk soils and were linked to distinct variations in bacterial functions across the soil-plant interface. Specifically, the ARGs enriched in the rhizosphere were significantly correlated to microbial functions of signaling pathways and degradation pathways. Conversely, the ARGs depleted in rhizosphere were significantly correlated to microbial functions of biosynthesis pathways, nutrient cycling, and ABC transport system. Moreover, our investigation involved metagenomic contig assembly, providing compelling evidence that these aforementioned relationships were indeed detected in the assembled bins.
Conclusions
The results of our study revealed a clear association between the distribution of ARGs and changes in bacterial functions attributable to the assemblage of the rhizosphere microbiome at the soil-plant interface. These findings significantly contribute to enhancing our comprehension of the mechanism underlying the transmission of ARGs from bulk soils to rhizosphere soils.







Similar content being viewed by others
References
Ai C, Liang G, Sun J, Wang X, He P, Zhou W, He X (2015) Reduced dependence of rhizosphere microbiome on plant-derived carbon in 32-year long-term inorganic and organic fertilized soils. Soil Biol Biochem 80:70–78
Andersson DI, Levin BR (1999) The biological cost of antibiotic resistance. Curr Opin Microbiol 2:489–493
Bai Y, Müller DB, Srinivas G, Garrido-Oter R, Schulze-Lefert P, Bai Y, Müller DB, Srinivas G, Garrido-Oter R, Potthoff E, Rott M, Dombrowski N, Münch PC, Spaepen S, Remus-Emsermann M, Hüttel B, McHardy AC, Vorholt JA, Schulze-Lefert P (2015) Functional overlap of the Arabidopsis leaf and root microbiota. Nature 528:364–369
Balsanelli E, Tadra-Sfeir MZ, Faoro H, Pankievicz VCS, de Baura VA, Pedrosa FO, de Souza EM, Dixon R, Monteiro RA (2016) Molecular adaptations of Herbaspirillum seropedicae during colonization of the maize rhizosphere. Environ Microbiol 18:2343–2356
Berendonk TU, Manaia CM, Merlin C, Fatta-Kassinos D, Cytryn E, Walsh F, Bürgmann H, Sørum H, Norström M, Pons M-N, Berendonk TU, Manaia CM, Merlin C, Fatta-Kassinos D, Cytryn E, Walsh F, Bürgmann H, Sørum H, Norström M, Pons M-N, Kreuzinger N, Huovinen P, Stefani S, Schwartz T, Kisand V, Baquero F, Martinez JL (2015) Tackling antibiotic resistance: the environmental framework. Nat Rev Microbiol 13:310–317
Bulgarelli D, Garrido-Oter R, Muench PC, Weiman A, Droege J, Pan Y, McHardy AC, Schulze-Lefert P (2015) Structure and function of the bacterial root microbiota in wild and domesticated barley. Cell Host Microbe 17:392–403
Carvalhais LC, Dennis PG, Badri DV, Kidd BN, Vivanco JM, Schenk PM (2015) Linking jasmonic acid signaling, root exudates, and rhizosphere microbiomes. Mol Plant Microbe in 28:1049–1058
Chen QL, An XL, Li H, Su J, Ma Y, Zhu YG (2016) Long-term field application of sewage sludge increases the abundance of antibiotic resistance genes in soil. Environ in 92–93:1–10
Chen QL, Cui HL, Su JQ, Penuelas J, Zhu YG (2019) Antibiotic resistomes in plant microbiomes. Trend Plant Sci 24:530–541
Choi CW, Yun SH, Kwon SO, Leem SH, Choi JS, Yun CY, Kim SI (2010) Analysis of cytoplasmic membrane proteome of streptococcus pneumoniae by shotgun proteomic approach. J Microbiol 48:872–876
de Vries FT, Wallenstein MD (2017) Below-ground connections underlying above-ground food production: a framework for optimizing ecological connections in the rhizosphere. J Ecol 105:913–920
Deng JM, Xiao XF, Fan WJ, Ning ZP, Xiao EZ (2022) relevance of the microbial community to Sb and as biogeochemical cycling in natural wetlands. Sci Tot Environ 818:151826
Doermann P, Kim H, Ott T, Schulze-Lefert P, Trujillo M, Wewer V, Hueckelhoven R (2014) Cell-autonomous defense, re-organization and trafficking of membranes in plant-microbe interactions. New Phytol 204:815–822
Donat S, Streker K, Schirmeister T, Rakette S, New Phytologist Stehle T, Liebeke M, Lalk M, Ohlsen K (2009) Transcriptome and functional analysis of the eukaryotic-type serine/threonine kinase PknB in Staphylococcus aureus. J Bacteriol 191:4056–4069
Edgell K (1989) USEPA method study 37 sw-846 method 3050 acid digestion of sediments, sludges, and soils. Report for September 1986-December 1987 (Final). US Environmental Protection Agency, Environmental Monitoring Systems Laboratory
Edwards J, Johnson C, Santos-Medellín C, Lurie E, Podishetty NK, Bhatnagar S, Eisen JA, Sundaresan V (2015) Structure, variation, and assembly of the root-associated microbiomes of rice. Proc Natl Acad Sci USA 112:911–920
El Khoury JY, Boucher N, Bergeron MG, Leprohon P, Ouellette M (2017) Penicillin induces alterations in glutamine metabolism in Streptococcus pneumoniae. Sci Rep 7:14587
Erickson MG, Ulijasz AT, Weisblum B (2005) Bacterial 2-component signal transduction systems: a fluorescence polarization screen for response regulator-protein binding. J Biomol Screen 10:270–274
Fahrenfeld N, Knowlton K, Krometis LA, Hession WC, Xia K, Lipscomb E, Libuit K, Green BL, Pruden A (2014) Effect of manure application on abundance of antibiotic resistance genes and their attenuation rates in soil: field-scale mass balance approach. Environ Sci Technol 48:2643–2650
Fan WJ, Deng JM, Shao L, Jiang SM, Xiao TF, Sun WM, Xiao EZ (2022) The rhizosphere microbiome improves the adaptive capabilities of plants under high soil cadmium conditions. Front Plant Sci 13:914103
Forsberg KJ, Patel S, Gibson MK, Lauber CL, Knight R, Fierer N, Dantas G (2014) Bacterial phylogeny structures soil resistomes across habitats. Nature 509:612–616
Gooderham WJ, Gellatly SL, Sanschagrin F, McPhee JB, Bains M, Cosseau C, Levesque RC, Hancock RE (2009) The sensor kinase PhoQ mediates virulence in Pseudomonas aeruginosa. Microbiology 155:699–711
Gustafson J, Strässle A, Hächler H, Kayser FH, Berger-Bächi B (1994) The femC locus of Staphylococcus aureus required for methicillin resistance includes the glutamine synthetase operon. J Bacteriol 176:1460–1467
Harth G, Horwitz MA (2003) Inhibition of Mycobacterium tuberculosis glutamine synthetase as a novel antibiotic strategy against tuberculosis: demonstration of efficacy in vivo. Infect Immun 71:456–464
Hong HJ, Hutchings MI, Neu JM, Wright GD, Paget MSB, Buttner MJ (2004) Characterization of an inducible vancomycin resistance system in Streptomyces coelicolor reveals a novel gene (vanK) required for drug resistance. Mol Microbiol 52:1107–1121
Hou Q, Kolodkin-Gal I (2020) Harvesting the complex pathways of antibiotic production and resistance of soil bacilli for optimizing plant microbiome. FEMS Microbiol Ecol 96:142
Hu HW, Wang JT, Li J, Shi XZ, Ma YB, Chen D, He JZ (2017) Long-term nickel contamination increases the occurrence of antibiotic resistance genes in agricultural soils. Environ Sci Technol 51:790–800
Jia B, Raphenya AR, Alcock B, Waglechner N, Guo P, Tsang KK, Lago BA, Dave BM, Pereira S, Sharma AN (2017) CARD 2017: expansion and model-centric curation of the comprehensive antibiotic resistance database. Nucleic Acids Res 45:566–573
Khan MT, Khan TA, Ahmad I, Muhammad S, Wei DQ (2022) Diversity and novel mutations in membrane transporters of Mycobacterium tuberculosis. Brief Funct Genomics 22:168–179
Leff JW, Jones SE, Prober SM, Barberán A, Borer ET, Firn JL, Harpole WS, Hobbie SE, Hofmockel KS, Knops JM (2015) Consistent responses of soil microbial communities to elevated nutrient inputs in grasslands across the globe. Proc Natl Acad Sci USA 112:10967–10972
Li X, Jousset A, de Boer W, Carrion VJ, Zhang T, Wang X, Kuramae EE (2019) Legacy of land use history determines reprogramming of plant physiology by soil microbiome. ISME J 13:738–751
Li Y, Yang R, Häggblom MM, Li M, Guo L, Li B, Kolton M, Cao Z, Soleimani M, Chen Z, Xu Z, Gao W, Yan B, Sun W (2022) Characterization of diazotrophic root endophytes in chinese silvergrass (Miscanthus sinensis). Microbiome 10(1):186
Lloyd NA, Nazaret S, Barkay T (2018) Whole genome sequences to assess the link between antibiotic and metal resistance in three coastal marine bacteria isolated from the mummichog gastrointestinal tract. Mar Pollut Bull 135:514–520
Long X, Ni W, Fu C, Yang X, Long X, Ni W, Fu C (2002) Sedum alfredii H: a new Zn hyperaccumulating plant first found in China. Chin Sci Bull 47:1634–1637
Love MI, Huber W, Anders S (2014) Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genom Biol 15:550
Luo G, Li B, Li LG, Zhang T, Angelidaki I (2017) Antibiotic resistance genes and correlations with microbial community and metal resistance genes in full-scale biogas reactors as revealed by metagenomic analysis. Environ Sci Technol 51:4069–4080
Mager DM, Thomas AD (2011) Extracellular polysaccharides from cyanobacterial soil crusts: a review of their role in dryland soil processes. J Arid Environ 75:91–97
Maria-Neto S, Cândido EdS, Rodrigues DR, de Sousa DA, da Silva EM, de Moraes LMP, Otero-Gonzalez AdJ, Magalhães BS, Dias SC, Franco OL (2012) Deciphering the magainin resistance process of Escherichia coli strains in light of the cytosolic proteome. Antimicrob Agents Ch 56:1714
Mende DR, Waller AS, Sunagawa S, Järvelin AI, Chan MM, Arumugam M, Raes J, Bork P (2012) Assessment of metagenomic assembly using simulated next generation sequencing data. Plos One 7:e31386
Merino-Martin L, Griffiths RI, Gweon HS, Furget-Bretagnon C, Oliver A, Mao Z, Le Bissonnais Y, Stokes A (2020) Rhizosphere bacteria are more strongly related to plant root traits than fungi in temperate montane forests: insights from closed and open forest patches along an elevational gradient. Plant Soil 450:183–200
Minoru K, Susumu G, Yoko S, Masayuki K, Miho F, Mao T (2014) Data, information, knowledge and principle: back to metabolism in KEGG. Nucleic Acids Res 42:199–205
Nielsen HB, Almeida M, Juncker AS, Rasmussen S, Li J, Sunagawa S, Plichta DR, Gautier L, Pedersen AG, Le Chatelier E, Pelletier E, Bonde I, Nielsen T, Nielsen HB, Almeida M, Juncker AS, Rasmussen S, Li J, Sunagawa S, Plichta DR, Gautier L, Pedersen AG, Le Chatelier E, Pelletier E, Bonde I, Nielsen T, Manichanh C, Arumugam M, Batto J-M, Quintanilha dos Santos MB, Blom N, Borruel N, Burgdorf KS, Boumezbeur F, Casellas F, Doré J, Dworzynski P, Guarner F, Hansen T, Hildebrand F, Kaas RS, Kennedy S, Kristiansen K, Kultima JR, Léonard P, Levenez F, Lund O, Moumen B, Le Paslier D, Pons N, Pedersen O, Prifti E, Qin J, Raes J, Sørensen S, Tap J, Tims S, Ussery DW, Yamada T, Renault P, Sicheritz-Ponten T, Bork P, Wang J, Brunak S, Ehrlich SD (2014) Identification and assembly of genomes and genetic elements in complex metagenomic samples without using reference genomes. Nat Biotechnol 32:822–828
Pedrosa FO, Monteiro RA, Wassem R, Cruz LM, Ayub RA, Colauto NB, Fernandez MA, Fungaro MH, Grisard EC, Hungria M, Madeira HM, Nodari RO, Osaku CC et al (2011) Genome of Herbaspirillum seropedicae strain SmR1, a specialized diazotrophic endophyte of tropical grasses. PLoS Genet 7:e1002064
Qi Q, Preston GM, MacLean RC (2014) Linking system-wide impacts of rna polymerase mutations to the fitness cost of rifampin resistance in Pseudomonas aeruginosa. MBio 5:e01562
Reynolds PE, Courvalin P (2005) Vancomycin resistance in enterococci due to synthesis of precursors terminating in alanyl serine. Antimicrob Agents Ch 49:21–25
Touati A, Bellil Z, Barache D, Mairi A (2021) Fitness cost of antibiotic resistance in Staphylococcus aureus: a systematic review. Microb Drug Resist 27:1218–1231
Udikovic-Kolic N, Wichmann F, Broderick NA, Handelsman J (2014) Bloom of resident antibiotic-resistant bacteria in soil following manure fertilization. Proc Natl Acad Sci USA 111:15202–15207
Wang C, Hu R, Strong PJ, Zhuang W, Huang W, Luo Z, Yan Q, He Z, Shu L (2021) Prevalence of antibiotic resistance genes and bacterial pathogens along the soil-mangrove root continuum. J Hazard Mater 408:124985
Wei H, Ding S, Qiao Z, Su Y, Xie B (2020) Insights into factors driving the transmission of antibiotic resistance from sludge compost-amended soil to vegetables under cadmium stress. Sci Tot Environ 729:138990
Weyrauch P, Heker I, Zaytsev AV, von Hagen CA, Arnold ME, Golding BT, Meckenstock RU (2020) The 5,6,7,8-Tetrahydro-2-naphthoyl-coenzyme A reductase reaction in the anaerobic degradation of naphthalene and identification of downstream metabolites. Appl Environ Microbiol 86:e00996–e00920
Wu Y, Simmons B, Singer S (2016) MaxBin 2.0: an automated binning algorithm to recover genomes from multiple metagenomic datasets. Bioinformatics 32:605–607
Xiao E, Ning Z, Xiao T, Sun W, Jiang S (2021) Soil bacterial community functions and distribution after mining disturbance. Soil Biology Biochem 157:108232
Xiao EZ, Sun WM, Ning ZP, Wang YQ, Meng FD, Deng JM, Fan WJ, Xiao TF (2022) Occurrence and dissemination of antibiotic resistance genes in mine soil ecosystems. Appl Microbiol Biotechnol 106:6289–6299
Xu Y, Xu J, Mao D, Luo Y (2017) Effect of the selective pressure of sub-lethal level of heavy metals on the fate and distribution of ARGs in the catchment scale. Environ Pollut 220:900–908
Xu J, Zhang Y, Zhang P, Trivedi P, Riera N, Wang Y, Liu X, Fan G, Tang J, Coletta-Filho HD, Cubero J, Deng X, Ancona V, Lu Z, Zhong B, Roper MC, Capote N, Catara V, Pietersen G, Vernière C, Al-Sadi AM, Li L, Yang F, Xu X, Wang J, Yang H, Jin T, Wang N (2018a) The structure and function of the global citrus rhizosphere microbiome. Nat Commun 9:4894
Xu X, Zarecki R, Medina S, Ofaim S, Liu X, Chen C, Hu S, Brom D, Gat D, Porob S, Eizenberg H, Ronen Z, Jiang J, Freilich S (2018b) Modeling microbial communities from atrazine contaminated soils promotes the development of biostimulation solutions. ISME J 13:494–508
Ye LM, Hildebrand F, Dingemans J, Ballet S, Laus G, Matthijs S, Berendsen R, Cornelis P (2014) Draft genome sequence analysis of a Pseudomonas putida W15Oct28 strain with antagonistic activity to Gram-positive and Pseudomonas sp pathogens. PLoS ONE 9:e110038
Yeung AT, Bains M, Hancock RE (2011) The sensor kinase CbrA is a global regulator that modulates metabolism, virulence, and antibiotic resistance in Pseudomonas aeruginosa. J Bacteriol 193:918–931
Zhang H, Chang F, Shi P, Ye L, Zhou Q, Pan Y, Li A (2019) Antibiotic Resistome Alteration by different disinfection strategies in a full-scale drinking Water Treatment Plant deciphered by Metagenomic Assembly. Environ Sci Technol 53:2141–2150
Zhang M, Chen L, Ye C, Yu X (2018) Co-selection of antibiotic resistance via copper shock loading on bacteria from a drinking water bio-filter. Environ Pollut 233:132–141
Acknowledgements
This research was funded by the National Natural Science Foundation of China (41807127, 41830753, U1612442) and the Foundation of Guangzhou City for basic research projects (202201010283, 202201010153).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no current or potential competing financial interests.
Additional information
Responsible Editor: Paul Bodelier.
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Xiao, E., Sun, W., Deng, J. et al. The potential linkage between antibiotic resistance genes and microbial functions across soil–plant systems. Plant Soil 493, 589–602 (2023). https://doi.org/10.1007/s11104-023-06247-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11104-023-06247-5