Abstract
Background and aims
Harnessing soil microbiomes is a major demand for development of sustainable and productive agriculture. Here we aimed to assess the impact of two different types of organic material amendments in combination with chemical fertilizer on the plant-soil microbiota in maize farming and its link to soil fertility and crop productivity.
Methods
Soils and roots were collected from a long-term wheat-maize rotation system involving three experimental treatments: chemical fertilizer (CF); chemical fertilizer plus seasonal application of manure (OM); and chemical fertilizer plus one-time application of peat and vermiculite (PV). Crop residues were returned in all three treatments each season. Bacterial 16S rRNA gene and fungal ITS sequencing were conducted to elucidate the treatment-specific response of the microbiota in bulk soil, rhizosphere soil, and root compartment.
Results
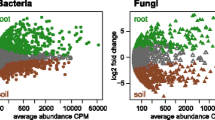
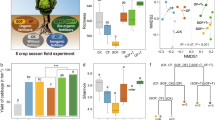
Relative to CF and OM treatments, PV amendment led to significant increases in soil organic carbon (SOC) content, aboveground plant biomass, and grain yield over the five-year field study. The PV-induced changes in microbial composition involved the greatest treatment-specific “effect size” on indicator ASVs (amplicon sequence variants) in bulk and rhizosphere soils. The number of interactions was more than doubled in the PV co-occurrence network relative to those in the CF and OM co-occurrence networks. Potential beneficial microbes, such as Glomeromycota (arbuscular mycorrhiza), Basidiomycota, and various members of the Actinobacteria and Burkholderiales, were most enriched in the root compartment of the PV treatment.
Conclusions
Peat-vermiculite enhanced microbiota-driven soil fertility and crop productivity, thereby providing new insights into plant-soil-microbiota interactions that can be harnessed for smart farming.





Similar content being viewed by others
References
Abarenkov K, Henrik NR, Larsson KH, Alexander IJ, Eberhardt U, Erland S, Kõljalg U (2010) The UNITE database for molecular identification of fungi - recent updates and future perspectives. New Phytol 186:281–285
Banerjee S, Walder F, Büchi L, Meyer M, Held AY, Gattinger A, Keller T, Charles R, van der Heijden MGA (2019) Agricultural intensification reduces microbial network complexity and the abundance of keystone taxa in roots. ISME J 13:1722–1736
Bei Q, Moser G, Wu X, Mueller C, Liesack W (2019) Metatranscriptomics reveals climate change effects on the rhizosphere microbiomes in European grassland. Soil Biol Biochem 138:107604
Bellemain E, Carlsen T, Brochmann C, Coissac E, Taberlet P, Kauserud H (2010) ITS as an environmental DNA barcode for fungi: an in-silico approach reveals potential PCR biases. BMC Microbiol 10:189
Bolger AM, Lohse M, Usadel B (2014) Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30:2114–2120
Busby PE, Soman C, Wagner MR, Friesen ML, Kremer J, Bennett A (2017) Research priorities for harnessing plant microbiomes in sustainable agriculture. PLoS Biol 15:e2001793
Chen X, Cui Z, Vitousek PM, Cassman KG, Matson PA, Bai JS, Meng QF, Hou P, Yue SC, Romheld V, Zhang FS (2011) Integrated soil-crop system management for food security. PNAS 108:6399–6404
Chen L, Brookes PC, Xu J, Zhang J, Zhang C, Zhou X, Luo Y (2016) Structural and functional differentiation of the root-associated bacterial microbiomes of perennial ryegrass. Soil Biol Biochem 98:1–10
Chen S, Waghmode TR, Sun R, Kuramae EE, Hu C, Liu B (2019) Root-associated microbiomes of wheat under the combined effect of plant development and nitrogen fertilization. Microbiome 7:136
Chen Q, Ding J, Zhu Y, He J, Hu H (2020) Soil bacterial taxonomic diversity is critical to maintaining the plant productivity. Environ Int 140:105766
de Vries FT, Wallenstein MD (2017) Below-ground connections underlying above ground food production: a framework for optimizing ecological connections in the rhizosphere. J Ecol 105:913–920
Deng Y, Wang S (2016) Synergistic growth in bacteria depends on substrate complexity. J Microbiol 54:23–30
Deru JGC, Bloem J, de Goede R, Hoekstra N, Keidel H, Kloen H, van Eekeren N (2019) Predicting soil N supply and yield parameters in peat grasslands. Applied Soil Ecol 134:77–84
Ding L, Cui H, Nie S, Long X, Duan G, Zhu Y (2019) Microbiomes inhabiting rice roots and rhizosphere. FEMS Microbiol Ecol 95
Edgar RC (2011) Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26:2460–2461
Edwards J, Johnson C, Santos-Medellín C, Lurie E, Podishetty NK, Bhatnagar S, Eisen JA, Sundaresan V (2015) Structure, variation, and assembly of the root-associated microbiomes of rice. PNAS 112:911–920
Fan K, Delgado-Baquerizo M, Guo X, Wang D, Zhu Y, Chu H (2020) Biodiversity of key-stone phylotypes determines crop production in a 4-decade fertilization experiment. ISME J
Fierer N (2017) Embracing the unknown: disentangling the complexities of the soil microbiome. Nat Rev Micro 15:579–590
Fitzpatrick CR, Copeland J, Wang PW, Guttman DS, Kotanen PM, Johnson MTJ (2018) Assembly and ecological function of the root microbiome across angiosperm plant species. PNAS 115:1157–1165
Gardeli C, Athenaki M, Xenopoulos E, Mallouchos AKAA, Aggelis G, Papanikolaou S (2017) Lipid production and characterization by Mortierella (Umbelopsis) isabellina cultivated on lignocellulosic sugars. J Appl Micro:1461–1477
Hale L, Luth M, Crowley D (2015) Biochar characteristics relate to its utility as an alternative soil inoculum carrier to peat and vermiculite. Soil Biol Biochem 81:228–235
Hartman K, van der Heijden MGA, Wittwer RA, Banerjee S, Walser J, Schlaeppi K (2018) Cropping practices manipulate abundance patterns of root and soil microbiome members paving the way to smart farming. Microbiome 6
Hermosilla E, Schalchli H, Mutis A, Diez MC (2017) Combined effect of enzyme inducers and nitrate on selective lignin degradation in wheat straw by Ganoderma lobatum. Environ Sci Pollut Res 24:21984–21996
Ho A, Lonardo DPD, Bodelier PLE (2017) Revisiting life strategy concepts in environmental microbial ecology. FEMS Microbiol Ecol 93:fix006
Hobbs PR, Sayre K, Gupta R (2008) The role of conservation agriculture in sustainable agriculture. Phil Trans R Soc B Biol Sci 363:543–555
Jeewani PH, Chen L, Van Zwieten L, Shen C, Guggenberger G, Luo Y, Xu J (2020) Shifts in the bacterial community along with root-associated compartments of maize as affected by goethite. Biol Fert Soils 56:1201–1210
Kandeler E, Stemmer M, Klimanek EM (1999) Response of soil microbial biomass, urease and xylanase within particle size fractions to long-term soil management. Soil Biol Biochem 31:261–273
Kielak AM, Cipriano MAP, Kuramae EE (2016) Acidobacteria strains from subdivision 1 act as plant growth-promoting bacteria. Arch Microbiol 198:987–993
Lal R (2004) Ecology: Managing Soil Carbon. Science 304:393
Lundell TK, Mäkelä MR, de Vries RP, Hildén KS (2014) Genomics, lifestyles and future prospects of wood-decay and litter-decomposing Basidiomycota. In fungi, Martin FM (ed) Advances in botanical research, London: Academic, pp 329-370
Ma L, Zhang W, Liu Z, Huang Y, Zhang Q, Tian X, Zhu Y (2020) Complete genome sequence of Streptomyces sp. SCSIO 03032 isolated from Indian Ocean sediment, producing diverse bioactive natural products. Mar Genom. https://doi.org/10.1016/j.margen.2020.100803
Magoč T, Steven L (2011) FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics 27:2957–2963
Martin M (2011) Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet 17:10–22
Martin F, Kohler A, Murat C, Veneault-Fourrey C, Hibbett DS (2016) Unearthing the roots of ectomycorrhizal symbioses. Nat Rev Microbiol 14:760–773
Niklasch H, Joergensen RG (2001) Decomposition of peat, biogenic municipal waste compost, and shrub/grass compost added in different rates to a silt loam. J Plant Nutr Soil Sci 164:365–369
Offre P, Pivato B, Mazurier S, Siblot S, Berta G, Lemanceau P (2008) Microdiversity of Burkholderiales associated with mycorrhizal and nonmycorrhizal roots of Medicago truncatula. FEMS Microbiol Ecol 65:180–192
Oksanen J, Kindt R, Legendre P, O'Hara B, Stevens MHH, Oksanen MJ, Suggests M (2007) The vegan package version 2.5-2. Community Ecol package. https://cran.r-project.org/web/packages/vegan
Öpik M, Vanatoa A, Vanatoa E, Moora M, Davison J, Kalwij JM, Reier Ü, Zobel M (2010) The online database MaarjAM reveals global and ecosystemic distribution patterns in arbuscular mycorrhizal fungi (Glomeromycota). New Phytol 188:223–241
Pan J, Shang Y, Zhang WJ, Chen X, Cui Z (2020) Improving soil quality for higher grain yields in Chinese wheat and maize production. Land Degrad Dev 31:1125–1137
Powell MJ (2016) Blastocladiomycota. In: Archibald J et al (eds) Handbook of the Protists. Springer, Cham. https://doi.org/10.1007/978-3-319-32669-6-17-1
Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, Glöckner FO (2012) The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res 41:590–596
Reganold JP, Wachter JM (2016) Organic agriculture in the twenty-first century. Nat Plants 2:15221
Robinson MD, McCarthy DJ, Smyth GK (2010) EdgeR: a bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26:139–140
Romano I, Ventorino V, Pepe O (2020) Effectiveness of plant beneficial microbes: overview of the methodological approaches for the assessment of root colonization and persistence. Front Plant Sci 11. https://doi.org/10.3389/fpls.2020.00006
Santoyo G, Moreno-Hagelsieb G, del Carmen O-MM, Glick BR (2016) Plant growth-promoting bacterial endophytes. Microbiol Res 183:92–99
Schmidt JE, Kent AD, Brisson VL, Gaudin ACM (2019) Agricultural management and plant selection interactively affect rhizosphere microbial community structure and nitrogen cycling. Microbiome 7:146
Schneijderberg M, Cheng X, Franken C, de Hollanderet M, van Velzen R, Schmitz L, Heinen R, Geurts R, van der Putten WH, T. Martijn Bezemer TM, Bisseling T (2020) Quantitative comparison between the rhizosphere effect of Arabidopsis thaliana and co-occurring plant species with a longer life history. ISME J https://doi.org/10.1038/s41396-020-0695-2
Shi S, Nuccio EE, Shi Z, He Z, Zhou J, Firestone MK (2016) The interconnected rhizosphere: high network complexity dominates rhizosphere assemblages. Ecol Lett 19:926–936
Smith SE, Read DJ (2008) Mycorrhizal symbiosis. Academic Press, Cambridge, UK
Tamaki H, Wright CL, Li X, Lin Q, Hwang C, Wang S, Thimmapuram J, Kamagata Y, Liu WT (2011) Analysis of 16S rRNA amplicon sequencing options on the Roche/454 next-generation titanium sequencing platform. PLoS One 6:e25263
Tedersoo L, Smith EM (2013) Lineages of ectomycorrhizal fungi revisited: foraging strategies and novel lineages revealed by sequences from belowground. Fungal Biol Rev 27:83–99
Toju H, Peay KG, Yamamichi M, Narisawa K, Hiruma K, Naito K, Fukuda S, Ushio M, Nakaoka S, Onoda Y, Yoshida K, Schlaeppi K, Bai Y, Sugiura R, Ichihashi Y, Minamisawa K, Kiers ET (2018) Core microbiomes for sustainable agroecosystems. Nat Plants 4:247–257
Trivedi P, Leach JE, Tringe SG, Sa T, Singh BK (2020) Plant–microbiome interactions: from community assembly to plant health. Nat Rev Microbiol 18:607–621. https://doi.org/10.1038/s41579-020-0412-1
van Bergeijk DA, Terlouw BR, Medema MH, van Wezel GP (2020) Ecology and genomics of Actinobacteria: new concepts for natural product discovery. Nat Rev Microbiol 18:546–558
van der Meij A, Worsley SF, Hutchings MI, van Wezel GP (2017) Chemical ecology of antibiotic production by actinomycetes. FEMS Microbiol Rev 41:392–416
Vasconcellos RLF, de Silva MCP, da Ribeiro CM, Cardoso EJBN (2010) Isolation and screening for plant growth-promoting (PGP) actinobacteria from Araucaria angustifolia rhizosphere soil. Sci Agric 67:743–746
Vejan P, Abdullah R, Khadiran T, Ismail S, Nasrulhaq Boyce A (2016) Role of plant growth promoting rhizobacteria in agricultural sustainability-a review. Molecules 21:573
Vieira S, Sikorski J, Dietz S, Herz K, Schrumpf M, Bruelheide H, Scheel D, Friedrich MW, Overmann J (2020) Drivers of the composition of active rhizosphere bacterial communities in temperate grasslands. ISME J 14:463–475
Wang R, Zhang H, Sun L, Qi G, Chen S, Zhao XY (2017) Microbial community composition is related to soil biological and chemical properties and bacterial wilt outbreak. Sci Rep 7:343
Wei Z, Gu Y, Friman V, Kowalchuk GA, Xu Y, Shen Q, Jousset A (2019) Initial soil microbiome composition and functioning predetermine future plant health. Sci Adv 5:w759
Xu H, Wang X, Li H, Yao H, Su J, Zhu YG (2014) Biochar impacts soil microbial community composition and nitrogen cycling in an acidic soil planted with rape. Environ Sci Technol 48:9391–9399
Xu Y, Wang T, Li H, Ren C, Chen J, Yang G, Wang X (2019) Variations of soil nitrogen-fixing microorganism communities and nitrogen fractions in a Robinia pseudoacacia chronosequence on the loess plateau of China. Catena 174:316–323
Zhu Y, Zhao Y, Zhu D, Gillings M, Penuelas J, Ok Y, Capon A, Banwart S (2019) Soil biota, antimicrobial resistance and planetary health. Environ Int 131:105059
Acknowledgements
This study was financially supported by the Project for the National Natural Science Foundation of China (41977038), China Agricultural University (2020TC050 & 2020TC144), and Beijing advanced discipline.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Responsible Editor: Yongguan Zhu
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
ESM 1
(DOCX 4404 kb)
Rights and permissions
About this article
Cite this article
Wu, X., Liu, Y., Shang, Y. et al. Peat-vermiculite alters microbiota composition towards increased soil fertility and crop productivity. Plant Soil 470, 21–34 (2022). https://doi.org/10.1007/s11104-021-04851-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11104-021-04851-x