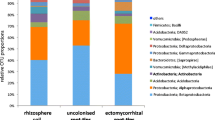
Abstract
Background and aims
Forest tree microbiomes are important to forest dynamics, diversity, and ecosystem processes. Mature limber pines (Pinus flexilis) host a core microbiome of acetic acid bacteria in their foliage, but the bacterial endophyte community structure, variation, and assembly across tree ontogeny is unknown. The aims of this study were to test if the core microbiome observed in adult P. flexilis is established at the seedling stage, if seedlings host different endophyte communities in root and shoot tissues, and how environmental factors structure seedling endophyte communities.
Methods
The 16S rRNA gene was sequenced to characterize the bacterial endophyte communities in roots and shoots of P. flexilis seedlings grown in plots at three elevations at Niwot Ridge, Colorado, subjected to experimental treatments (watering and heating). The data was compared to previously sequenced endophyte communities from adult tree foliage sampled in the same year and location.
Results
Seedling shoots hosted a different core microbiome than adult tree foliage and were dominated by a few OTUs in the family Oxalobacteraceae, identical or closely related to strains with antifungal activity. Shoot and root communities significantly differed from each other but shared major OTUs. Watering but not warming restructured the seedling endophyte communities.
Conclusions
The results suggest differences in assembly and ecological function across conifer life stages. Seedlings may recruit endophytes to protect against fungi under increased soil moisture.





Similar content being viewed by others
References
Altschul SF, Gish W, Miller W et al (1990) Basic local alignment search tool. J Mol Biol 215:403–410. https://doi.org/10.1016/S0022-2836(05)80360-2
Anand R, Grayston S, Chanway C (2013) N2-fixation and seedling growth promotion of lodgepole pine by endophytic Paenibacillus polymyxa. Microb Ecol 66:369–374. https://doi.org/10.1007/s00248-013-0196-1
Aronesty E (2011) Ea-utils : “Command-line tools for processing biological sequencing data”. https://github.com/ExpressionAnalysis/ea-utils
Bal A, Anand R, Berge O, Chanway C (2012) Isolation and identification of diazotrophic bacteria from internal tissues of Pinus contorta and Thuja plicata. Can J For Res 813:807–813. https://doi.org/10.1139/x2012-023
Baldani JI, Rouws L, Cruz LM et al (2014) The family Oxalobacteraceae. In: Rosenberg E, DeLong EF, Lory S et al (eds) The prokaryotes. Springer Berlin Heidelberg, pp 919–974
Beckers B, De Beeck MO, Weyens N et al (2017) Structural variability and niche differentiation in the rhizosphere and endosphere bacterial microbiome of field-grown poplar trees. Microbiome 5:1–17. https://doi.org/10.1186/s40168-017-0241-2
Bell CW, Tissue DT, Loik ME et al (2014) Soil microbial and nutrient responses to 7 years of seasonally altered precipitation in a Chihuahuan Desert grassland. Glob Chang Biol 20:1657–1673. https://doi.org/10.1111/gcb.12418
Bi J, Zhang N, Liang Y et al (2012) Interactive effects of water and nitrogen addition on soil microbial communities in a semiarid steppe. J Plant Ecol 5:320–329. https://doi.org/10.1093/jpe/rtr046
Bodenhausen N, Horton MW, Bergelson J (2013) Bacterial communities associated with the leaves and the roots of Arabidopsis thaliana. PLoS One 8:e56329. https://doi.org/10.1371/journal.pone.0056329
Bokulich NA, Subramanian S, Faith JJ et al (2013) Quality-filtering vastly improves diversity estimates from Illumina amplicon sequencing. Nat Methods 10:57–59. https://doi.org/10.1038/nmeth.2276
Bonito G, Reynolds H, Robeson MS et al (2014) Plant host and soil origin influence fungal and bacterial assemblages in the roots of woody plants. Mol Ecol 23:3356–3370. https://doi.org/10.1111/mec.12821
Brooks DD, Chan R, Starks ER et al (2011) Ectomycorrhizal hyphae structure components of the soil bacterial community for decreased phosphatase production. FEMS Microbiol Ecol 76:245–255. https://doi.org/10.1111/j.1574-6941.2011.01060.x
Bulgarelli D, Rott M, Schlaeppi K et al (2012) Revealing structure and assembly cues for Arabidopsis root-inhabiting bacterial microbiota. Nature 488:91–95. https://doi.org/10.1038/nature11336
Bulgari D, Bozkurt AI, Casati P et al (2012) Endophytic bacterial community living in roots of healthy and “Candidatus Phytoplasma mali”-infected apple (Malus domestica, Borkh.) trees. Antonie van Leeuwenhoek, Int J Gen Mol Microbiol 102:677–687. https://doi.org/10.1007/s10482-012-9766-3
Caporaso JG, Bittinger K, Bushman FD et al (2010a) PyNAST: a flexible tool for aligning sequences to a template alignment. Bioinformatics 26:266–267. https://doi.org/10.1093/bioinformatics/btp636
Caporaso JG, Kuczynski J, Stombaugh J et al (2010b) QIIME allows analysis of high- throughput community sequencing data. Nat Publ Group 7:335–336. https://doi.org/10.1038/nmeth0510-335
Carrell AA, Frank AC (2014) Pinus flexilis and Picea engelmannii share a simple and consistent needle endophyte microbiota with a potential role in nitrogen fixation. Front Microbiol 5:1–11. https://doi.org/10.3389/fmicb.2014.00333
Carrell AA, Frank AC (2015) Bacterial endophyte communities in the foliage of coast redwood and giant sequoia. Front Microbiol 6:1–11. https://doi.org/10.3389/fmicb.2015.01008
Carrell AA, Carper DL, Frank AC (2016) Subalpine conifers in different geographical locations host highly similar foliar bacterial endophyte communities. FEMS Microbiol Ecol 92:1–9. https://doi.org/10.1093/femsec/fiw124
Castanha C, Torn MS, Germino MJ et al (2013) Conifer seedling recruitment across a gradient from forest to alpine tundra: effects of species, provenance, and site. Plant Ecolog Divers 6:307–318. https://doi.org/10.1080/17550874.2012.716087
Castrillo G, Teixeira PJPL, Paredes SH et al (2017) Root microbiota drive direct integration of phosphate stress and immunity. Nature 543:513–518. https://doi.org/10.1038/nature21417
Chelius MK, Triplett EW (2001) The diversity of archaea and bacteria in association with the roots of Zea mays L. Microb Ecol 41:252–263. https://doi.org/10.1007/s002480000087
Coleman-Derr D, Desgarennes D, Fonseca-Garcia C et al (2016) Plant compartment and biogeography affect microbiome composition in cultivated and native Agave species. New Phytol 209:798–811. https://doi.org/10.1111/nph.13697
DeAngelis KM, Pold G, Topçuoglu BD et al (2015) Long-term forest soil warming alters microbial communities in temperate forest soils. Front Microbiol 6:1–13. https://doi.org/10.3389/fmicb.2015.00104
DeLeon-Rodriguez N, Lathem TL, Rodriguez-R LM et al (2013) Microbiome of the upper troposphere: species composition and prevalence, effects of tropical storms, and atmospheric implications. Proc Natl Acad Sci U S A 110:2575–2580. https://doi.org/10.1073/pnas.1212089110
Edgar RC (2013) UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat Methods 10:996–998. https://doi.org/10.1038/nmeth.2604
Edwards J, Johnson C, Santos-Medellín C et al (2015) Structure, variation, and assembly of the root-associated microbiomes of rice. Proc Natl Acad Sci U S A 112:E911–E920. https://doi.org/10.1073/pnas.1414592112
Elbeltagy A, Nishioka K, Sato T et al (2001) Endophytic colonization and in planta nitrogen fixation by a Herbaspirillum sp . isolated from wild rice species. Appl Environ Microbiol 67:5285–5293. https://doi.org/10.1128/AEM.67.11.5285
Fesel PH, Zuccaro A (2016) Dissecting endophytic lifestyle along the parasitism/mutualism continuum in Arabidopsis. Curr Opin Microbiol 32:103–112. https://doi.org/10.1016/j.mib.2016.05.008
García-Echauri SA, Gidekel M, Gutiérrez-Moraga A et al (2011) Isolation and phylogenetic classification of culturable psychrophilic prokaryotes from the Collins glacier in the Antarctica. Folia Microbiol (Praha) 56:209–214. https://doi.org/10.1007/s12223-011-0038-9
García-Salamanca A, Molina-Henares MA, van Dillewijn P et al (2013) Bacterial diversity in the rhizosphere of maize and the surrounding carbonate-rich bulk soil. Microb Biotechnol 6:36–44. https://doi.org/10.1111/j.1751-7915.2012.00358.x
Germino MJ, Smith WK (1999) Sky exposure, crown architecture, and low-temperature photoinhibition in conifer seedlings at alpine treeline. Plant Cell Environ 22:407–415. https://doi.org/10.1046/j.1365-3040.1999.00426.x
Germino MJ, Smith WK, Resor AC (2002) Conifer seedling distribution and survival in an alpine-treeline ecotone. Plant Ecol 162:157–168. https://doi.org/10.1023/A:1020385320738
Gottel NR, Castro HF, Kerley M et al (2011) Distinct microbial communities within the endosphere and rhizosphere of Populus deltoides roots across contrasting soil types. Appl Environ Microbiol 77:5934–5944. https://doi.org/10.1128/AEM.05255-11
Green SJ, Michel FC, Hadar Y, Minz D (2007) Contrasting patterns of seed and root colonization by bacteria from the genus Chryseobacterium and from the family Oxalobacteraceae. ISME J 1:291–299. https://doi.org/10.1038/ismej.2007.33
Gundale MJ, Kardol P, Nilsson MC et al (2014) Interactions with soil biota shift from negative to positive when a tree species is moved outside its native range. New Phytol 202:415–421. https://doi.org/10.1111/nph.12699
Haack FS, Poehlein A, Kroger C et al (2016) Molecular keys to the Janthinobacterium and Duganella spp. interaction with the plant pathogen Fusarium graminearum. Front Microbiol 7. https://doi.org/10.3389/fmicb.2016.01668
He L, Zhang Z, Zhong X, Sheng X (2017) Isolation and characterization of Rumex acetosa -associated mineral-weathering bacteria. Geomicrobiol J 0:1–8. https://doi.org/10.1080/01490451.2017.1338800
Hunziker L, Bönisch D, Groenhagen U et al (2015) Pseudomonas strains naturally associated with potato plants produce volatiles with high potential for inhibition of Phytophthora infestans. Appl Environ Microbiol 81:821–830. https://doi.org/10.1128/AEM.02999-14
Ikeda S, Sasaki K, Okubo T et al (2014) Low nitrogen fertilization adapts rice root microbiome to low nutrient rnvironment by changing biogeochemical functions. Microbes Environ 29:50–59. https://doi.org/10.1264/jsme2.ME13110
Jiao JY, Wang HX, Zeng Y, Shen YM (2006) Enrichment for microbes living in association with plant tissues. J Appl Microbiol 100:830–837. https://doi.org/10.1111/j.1365-2672.2006.02830.x
Jones SE, Newton RJ, McMahon KD (2008) Potential for atmospheric deposition of bacteria to influence bacterioplankton communities. FEMS Microbiol Ecol 64:388–394. https://doi.org/10.1111/j.1574-6941.2008.00476.x
Kloepper JW, Ryu CM (2006) Bacterial endophytes as elicitors of induced systemic resistance. In: B.J.E. S, Boyle CJ., Sieber TN (eds) Microbial Root Endophytes. Soil Biology, vol 9. Springer Berlin, Heidelberg, pp 33–52
Kost T, Stopnisek N, Agnoli K et al (2013) Oxalotrophy, a widespread trait of plant-associated Burkholderia species, is involved in successful root colonization of lupin and maize by Burkholderia phytofirmans. Front Microbiol 4:1–9. https://doi.org/10.3389/fmicb.2013.00421
Kueneman JG, Woodhams DC, Harris R et al (2016) Probiotic treatment restores protection against lethal fungal infection lost during amphibian captivity. Proc R Soc B Biol Sci 283:20161553. https://doi.org/10.1098/rspb.2016.1553
Kueppers LM, Conlisk E, Castanha C et al (2017a) Warming and provenance limit tree recruitment across and beyond the elevation range of subalpine forest. Glob Chang Biol 23:2383–2395. https://doi.org/10.1111/gcb.13561
Kueppers LM, Faist A, Ferrenberg S et al (2017b) Lab and field warming similarly advance germination date and limit germination rate for high and low elevation provenances of two widespread subalpine conifers. Forests 8:1–17. https://doi.org/10.3390/f8110433
La Porta N, Capretti P, Thomsen IM et al (2008) Forest pathogens with higher damage potential due to climate change in Europe. Can J Plant Pathol 30:177–195. https://doi.org/10.1080/07060661.2008.10540534
Larkin BG, Hunt LS, Ramsey PW (2012) Foliar nutrients shape fungal endophyte communities in western white pine (Pinus monticola) with implications for white-tailed deer herbivory. Fungal Ecol 5:252–260. https://doi.org/10.1016/j.funeco.2011.11.002
Lebels SI, Paredes SH, Lundberg DS et al (2015) Salicylic acid modulates colonization of the root microbiome by specific bacterial taxa. Science (80- ) 349:860–864. https://doi.org/10.5061/dryad.238b2
Lee YM, Kim EH, Lee HK, Hong SG (2014) Biodiversity and physiological characteristics of Antarctic and Arctic lichens-associated bacteria. World J Microbiol Biotechnol 30:2711–2721. https://doi.org/10.1007/s11274-014-1695-z
Leveau JHJ, Uroz S, de Boer W (2010) The bacterial genus Collimonas: mycophagy, weathering and other adaptive solutions to life in oligotrophic soil environments. Environ Microbiol 12:281–292. https://doi.org/10.1111/j.1462-2920.2009.02010.x
Liu Q, Liu HC, Zhang JL et al (2015) Sphingomonas psychrolutea sp. nov., a psychrotolerant bacterium isolated from glacier ice. Int J Syst Evol Microbiol 65:2955–2959. https://doi.org/10.1099/ijs.0.000362
Lòpez-Fernàndez S, Mazzoni V, Pedrazzoli F et al (2017) A phloem-feeding insect transfers bacterial endophytic communities between grapevine plants. Front Microbiol 8:1–17. https://doi.org/10.3389/fmicb.2017.00834
López-Mondéjar R, Zühlke D, Becher D et al (2016) Cellulose and hemicellulose decomposition by forest soil bacteria proceeds by the action of structurally variable enzymatic systems. Sci Rep 6:25279. https://doi.org/10.1038/srep25279
Lozupone C, Knight R (2005) UniFrac : a new phylogenetic method for comparing microbial communities. Appl Environ Microbiol 71:8228–8235. https://doi.org/10.1128/AEM.71.12.8228
Lundberg DS, Lebeis SL, Paredes SH et al (2012) Defining the core Arabidopsis thaliana root microbiome. Nature 488:86–90. https://doi.org/10.1038/nature11237
Marasco R, Rolli E, Ettoumi B et al (2012) A drought resistance-promoting microbiome is selected by root system under desert farming. PLoS One 7:e48479. https://doi.org/10.1371/journal.pone.0048479
McMurdie PJ, Holmes S (2013) Phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One 8:e61217. https://doi.org/10.1371/journal.pone.0061217
Mishra A, Gond SK, Kumar A et al (2012) Season and tissue type affect fungal endophyte communities of the Indian medicinal plant Tinospora cordifolia more strongly than geographic location. Microb Ecol 64:388–398. https://doi.org/10.1007/s00248-012-0029-7
Moyes AB, Castanha C, Germino MJ, Kueppers LM (2013) Warming and the dependence of limber pine (Pinus flexilis) establishment on summer soil moisture within and above its current elevation range. Oecologia 171:271–282. https://doi.org/10.1007/s00442-012-2410-0
Moyes AB, Kueppers LM, Pett-Ridge J et al (2016) Evidence for foliar endophytic nitrogen fixation in a widely distributed subalpine conifer. New Phytol 210:657–668. https://doi.org/10.1111/nph.13850
Murakami T, Segawa T, Bodington D et al (2015) Census of bacterial microbiota associated with the glacier ice worm Mesenchytraeus solifugus. FEMS Microbiol Ecol 91:1–10. https://doi.org/10.1093/femsec/fiv003
Naveed M, Mitter B, Reichenauer TG et al (2014) Increased drought stress resilience of maize through endophytic colonization by Burkholderia phytofirmans PsJN and Enterobacter sp. FD17. Environ Exp Bot 97:30–39. https://doi.org/10.1016/j.envexpbot.2013.09.014
Nissinen RM, Männistö MK, van Elsas JD (2012) Endophytic bacterial communities in three arctic plants from low arctic fell tundra are cold-adapted and host-plant specific. FEMS Microbiol Ecol 82:510–522. https://doi.org/10.1111/j.1574-6941.2012.01464.x
Ofek M, Hadar Y, Minz D (2012) Ecology of root colonizing Massilia (Oxalobacteraceae). PLoS One 7:e40117. https://doi.org/10.1371/journal.pone.0040117
Oksanen J, Blanchet FG, Friendly M et al (2018) Vegan: Community Ecology Package. R package version 2.5-1. https://CRAN.R-project.org/package=vegan
Ondov BD, Bergman NH, Phillippy AM (2011) Interactive metagenomic visualization in a web browser. BMC Bioinformatics 12:385. https://doi.org/10.1186/1471-2105-12-385
Oono R, Lefèvre E, Simha A, Lutzoni F (2015) A comparison of the community diversity of foliar fungal endophytes between seedling and adult loblolly pines (Pinus taeda). Fungal Biol 119:917–928. https://doi.org/10.1016/j.funbio.2015.07.003
Oteino N, Lally RD, Kiwanuka S et al (2015) Plant growth promotion induced by phosphate solubilizing endophytic Pseudomonas isolates. Front Microbiol 6:1–9. https://doi.org/10.3389/fmicb.2015.00745
Paiva G, Abreu P, Neves Proença D et al (2014) Mucilaginibacter pineti sp. nov., isolated from Pinus pinaster wood from a mixed grove of pines trees. Int J Syst Evol Microbiol 64:2223–2228. https://doi.org/10.1099/ijs.0.057737-0
Paulson JN, Stine OC, Bravo HC, Pop M (2013) Differential abundance analysis for microbial marker-gene surveys. Nat Methods 10:1200–1202. https://doi.org/10.1038/nmeth.2658
Paulson JN, Talukder H, Pop M, Bravo HC (2016) metagenomeSeq: Statistical analysis for sparse high-throughput sequencing. Bioconductor package: 1.20.1. http://cbcb.umd.edu/software/metagenomeSeq
Peiffer JA, Spor A, Koren O et al (2013) Diversity and heritability of the maize rhizosphere microbiome under field conditions. Proc Natl Acad Sci 110:6548–6553. https://doi.org/10.1073/pnas.1302837110
Poosakkannu A, Nissinen R, Kytöviita MM (2015) Culturable endophytic microbial communities in the circumpolar grass, Deschampsia flexuosa in a sub-Arctic inland primary succession are habitat and growth stage specific. Environ Microbiol Rep 7:111–122. https://doi.org/10.1111/1758-2229.12195
Price MN, Dehal PS, Arkin AP (2009) Fasttree: computing large minimum evolution trees with profiles instead of a distance matrix. Mol Biol Evol 26:1641–1650. https://doi.org/10.1093/molbev/msp077
Pruesse E, Quast C, Knittel K et al (2007) SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res 35:7188–7196. https://doi.org/10.1093/nar/gkm864
Redford AJ, Bowers RM, Knight R et al (2010) The ecology of the phyllosphere: geographic and phylogenetic variability in the distribution of bacteria on tree leaves. Environ Microbiol 12:2885–2893. https://doi.org/10.1111/j.1462-2920.2010.02258.x.The
Reinhardt K, Castanha C, Germino MJ, Kueppers LM (2011) Ecophysiological variation in two provenances of Pinus flexilis seedlings across an elevation gradient from forest to alpine. Tree Physiol 31:615–625. https://doi.org/10.1093/treephys/tpr055
Robinson RJ, Fraaije BA, Clark IM et al (2016) Endophytic bacterial community composition in wheat (Triticum aestivum) is determined by plant tissue type, developmental stage and soil nutrient availability. Plant Soil 405:381–396. https://doi.org/10.1007/s11104-015-2495-4
Rodriguez RJ, White JF, Arnold a E, Redman RS (2009) Fungal endophytes: diversity and functional roles. New Phytol 182:314–330. https://doi.org/10.1111/j.1469-8137.2009.02773.x
Santos-Medellín C, Edwards J, Liechty Z et al (2017) Drought stress results in a compartment-specific restructuring of the rice root-associated microbiomes. MBio 8:e00764–e00717
Schindlbacher A, Rodler A, Kuffner M et al (2011) Experimental warming effects on the microbial community of a temperate mountain forest soil. Soil Biol Biochem 43:1417–1425. https://doi.org/10.1016/j.soilbio.2011.03.005
Schlaeppi K, Bulgarelli D (2015) The plant microbiome at work. Mol Plant-Microbe Interact 28:212–217. https://doi.org/10.1094/MPMI-10-14-0334-FI
Schlaeppi K, Dombrowski N, Oter RG et al (2014) Quantitative divergence of the bacterial root microbiota in Arabidopsis thaliana relatives. Proc Natl Acad Sci U S A 111:585–592. https://doi.org/10.1073/pnas.1321597111
Shakya M, Gottel N, Castro H et al (2013) A multifactor analysis of fungal and bacterial community structure in the root microbiome of mature Populus deltoides trees. PLoS One 8:e76382. https://doi.org/10.1371/journal.pone.0076382
Shen SY, Fulthorpe R (2015) Seasonal variation of bacterial endophytes in urban trees. Front Microbiol 6:1–13. https://doi.org/10.3389/fmicb.2015.00427
Smith WK, Germino MJ, Hancock TE, Johnson DM (2003) Another perspective on altitudinal limits of alpine timberlines. Tree Physiol 23:1101–1112. https://doi.org/10.1093/treephys/23.16.1101
Stinson M, Hess WM, Sears J, Strobel G (2003) An endophytic Gliocladium sp . of Eucryphia cordifolia producing selective volatile antimicrobial compounds. Plant Sci 165:913–922. https://doi.org/10.1016/S0168-9452(03)00299-1
Sziderics AH, Rasche F, Trognitz F et al (2007) Bacterial endophytes contribute to abiotic stress adaptation in pepper plants (Capsicum annuum L.). Can J Microbiol 53:1195–1202. https://doi.org/10.1139/W07-082
Vasiliauskas R, Menkis A, Finlay RD, Stenlid J (2007) Wood-decay fungi in fine living roots of conifer seedlings. New Phytol 174:441–446. https://doi.org/10.1111/j.1469-8137.2007.02014.x
Venables W, Ripley B (2002) Modern applied statistics with S, Fourth Edi. Springer, New York
Vences M, Granzow S, Künzel S et al (2015) Composition and variation of the skin microbiota in sympatric species of European newts (Salamandridae). Amphibia-Reptilia 36:5–12. https://doi.org/10.1163/15685381-00002970
Wagner MR, Lundberg DS, del Rio TG et al (2016) Host genotype and age shape the leaf and root microbiomes of a wild perennial plant. Nat Commun 7:1–15. https://doi.org/10.1038/ncomms12151
Walke JB, Becker MH, Hughey MC et al (2015) Most of the dominant members of amphibian skin bacterial communities can be readily cultured. Appl Environ Microbiol 81:6589–6600. https://doi.org/10.1128/AEM.01486-15
Wang Y, Naumann U, Wright ST, Warton DI (2012) Mvabund- an R package for model-based analysis of multivariate abundance data. Methods Ecol Evol 3:471–474. https://doi.org/10.1111/j.2041-210X.2012.00190.x
War AR, Paulraj MG, Ahmad T et al (2012) Mechanisms of plant defense against insect herbivores. Plant Signal Behav 7:1306–1320. https://doi.org/10.4161/psb.21663
Warton DI, Wright ST, Wang Y (2012) Distance-based multivariate analyses confound location and dispersion effects. Methods Ecol Evol 3:89–101. https://doi.org/10.1111/j.2041-210X.2011.00127.x
Xie Y, Wang X, Silander JA (2015) Deciduous forest responses to temperature, precipitation, and drought imply complex climate change impacts. Proc Natl Acad Sci 112:13585–13590. https://doi.org/10.1073/pnas.1509991112
Yao M, Rui J, Niu H et al (2017) The differentiation of soil bacterial communities along a precipitation and temperature gradient in the eastern inner Mongolia steppe. Catena 152:47–56. https://doi.org/10.1016/j.catena.2017.01.007
Yeoh YK, Dennis PG, Paungfoo-Lonhienne C et al (2017) Evolutionary conservation of a core root microbiome across plant phyla along a tropical soil chronosequence. Nat Commun 8:215. https://doi.org/10.1038/s41467-017-00262-8
Zhang X, Zhang G, Chen Q, Han X (2013) Soil bacterial communities respond to climate changes in a temperate steppe. PLoS One 8:1–9. https://doi.org/10.1371/journal.pone.0078616
Acknowledgements
This research was supported by National Science Foundation grant DEB-1442348 to ACF and LMK, and by an Office of Science (BER), US Department of Energy grant to LMK. We thank the Mountain Research Station and Niwot Ridge LTER at the University of Colorado, Boulder, for logistical support. Thanks to E. Brown, C. Castanha for field assistance. The sequencing was carried out by the DNA Technologies and Expression Analysis Cores at the UC Davis Genome Center, supported by NIH Shared Instrumentation Grant 1S10OD010786-01.
Author information
Authors and Affiliations
Corresponding author
Additional information
Responsible Editor: Stéphane Compant.
Electronic supplementary material
Fig. S1
Rarefaction curve showing shoot- and root samples. There is no apparent asymptote in the rarefaction curve suggesting that the community was undersampled. Lower numbers of OTUs were recovered from shoots than roots. The error bars represent standard error. (GIF 25 kb)
Rights and permissions
About this article
Cite this article
Carper, D.L., Carrell, A.A., Kueppers, L.M. et al. Bacterial endophyte communities in Pinus flexilis are structured by host age, tissue type, and environmental factors. Plant Soil 428, 335–352 (2018). https://doi.org/10.1007/s11104-018-3682-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11104-018-3682-x