Abstract
Purpose
Roots are inhabited by a broad range of fungi, including pathogens and mycorrhizal fungi, with functional traits related to plant health and nutrition. Management of these fungi in agroecosystems requires profound knowledge about their ecology. The main objective of this study was to examine succession patterns of root-associated fungi in pea during a full plant growth cycle.
Methods
Plants were grown in pots with field soil in a growth chamber under controlled conditions. Fungal communities in pea roots were analyzed at different plant growth stages including the vegetative growth, flowering and senescence, using 454 pyrosequencing.
Results
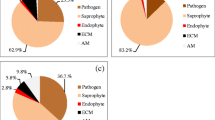
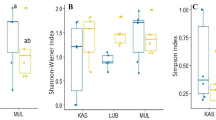
One hundred and twenty one non-singleton operational taxonomic units (OTUs) representing fungal species were detected. Pathogenic and arbuscular mycorrhizal fungi dominated during the vegetative growth stage, whereas saprotrophic fungi dominated during plant senescence.
Conclusions
In conclusion, the results from the present study demonstrated highly diverse fungal communities in pea roots with clear succession patterns related to fungal traits.


Similar content being viewed by others
References
Acosta-Martínez V, Dowd S, Sun Y, Allen V (2008) Tag-enhanced pyrosequencing analysis of bacterial diversity in a single soil type as affected by management and land use. Soil Biol Biochem 40:2562–2570
Bellemain E, Carlsen T, Brochmann C, Coissac E, Taberlet P, Kauserud H (2010) ITS as an environmental DNA barcode for fungi: an in silico approach reveals potential PCR biases. BMC Microbiol 10:189
Blackwell M (2011) The Fungi: 1, 2, 3, … 5, 1 million species? Biodiversity special issue. Am J Bot 98:426–438
Buée M, Reich M, Murat C, Morin E, Nilsson RH, Uroz S, Martin F (2009) 454 pyrosequencing analyses of forest soils reveal on unexpectedly high fungal diversity. New Phytol 184:449–456
Colwell RK (2009) EstimateS: Statistical estimation of species richness and shared species from samples. Version 8.2. User’s Guide and application. http://purl.oclc.org/estimates
Eissenstat DM, Yanai RD (2002) Root life span, efficiency, and turnover. In: Waisel Y, Eshel S, Kafkafi U (eds) Plant root: the hidden half. Marcel Dekker, New York, pp 221–238
Gardes M, Bruns TD (1993) ITS primers with enhanced specificity for basidiomycetes application to the identification of mycorrhizal and rusts. Mol Ecol 2:113–118
Gianinazzi S, Trouvelot A, Lovato P, Tuinen DV, Franken P, Gianinazzi-Pearson V (1995) Arbuscular Mycorrhizal Fungi in Plant Production of Temperate Agroecosystems. Crit Rev Biotech 15:305–311
Giovannetti M, Mosse B (1980) An evaluation of techniques for measuring vesicular arbuscular mycorrhizal infection in roots. New Phytol 84:489–500
Gransee A, Wittenmayer L (2000) Qualitative and quantitative analysis of water-soluble root exudates in relation to plant species and development. J Plant Nutr Soil Sci 163:381–385
Houlden A, Timms-Wilson TM, Day MJ, Bailey MJ (2008) Influence of plant developmental stage on microbial community structure and activity in the rhizosphere of three field crops. FEMS Microbiol Ecol 65:193–201
Hui N, Jumpponen A, Niskanen T, Liimatainen K, Jones KL, Koivula T (2011) EcM fungal community structure, but not diversity, altered in a Pb-contaminated shooting range in a boreal coniferous forest site in Southern Finland. FEMS Microbiol Ecol 76:121–132
Jumpponen A, Jones KL, Mattox JD, Yaege C (2010) Massively parallel in Quercus spp. ectomycorrhizas indicates seasonal dynamics in urban and rural sites. Mol Ecol 19:41–53
Kjøller R, Rosendahl S (2001) Molecular diversity of glomalean (arbuscular mycorrhizal) fungi determined as distinct Glomus specific DNA sequences from roots of field grown peas. Mycol Res 105:1027–1032
Kormanik PP, McGraw AC (1982) Quantification of vesicular-arbuscular mycorrhiza in plant roots. In: Schenck NC (ed) Methods and principles of mycorrhizal research. American Phytopathological Society, St. Paul, pp 37–45
Kraft JM, Pfleger FL (2001) Compendium of pea diseases and pests. In: Kraft JM, Pfleger FL (eds) The disease compendium series of the American Phytopathological Society. Am Phytopathol Soci, St. Paul
Kuzyakov Y, Domanski G (2000) Carbon input by plants into the soil. J Plant Nutr Soil Sci 163:421–431
Larsen J, Bødker L (2002) Interactions between pea root-associated fungi examined using signature fatty acids. New Phytol 149:487–493
Lindahl BD, Ihrmark K, Boberg J, Trumbore SE, Högberg P, Stenlid J, Finlay RD (2007) Spatial separation of litter decomposition and mycorrhizal nitrogen uptake in a boreal forest. New Phythol 173:611–620
Lumini E, Orgiazzi A, Borriello R, Bonfante P, Bianciotto V (2010) Disclosing arbuscualr mycorrhizal fungal biodiversity in soil through a land-use gradient using a pyrosequencing approach. Environ Microbiol 12:2165–2179
Margulies M, Egholm M, Altman WE, Attiya S et al (2005) Genome sequencing in microfabricated high-density picolitre reactors. Nature 437:376–380
Martin KJ, Rygiewicz PT (2005) Fungal-specific PCR primers developed for analysis of the ITS region of environmental DNA extracts. BMC Microbiol 5:28
Monchy S, Sanciu G, Jobard M, Rasconi S, Gerphagnon M, Chabé M, Cian A, Meloni D, Niquil N, Christaki U, Viscogliosi E, Sime-Ngando T (2011) Exploring and quantifying fungal diversity in freshwater lake ecosystems using rDNA cloning/sequencing and SSU tag pyrosequencing. Environ Microbiol 13:1433–1453
O’Brien HE, Parrent JL, Jackson JA, Moncalvo JM, Vilgalys R (2005) Fungal community analysis by large-scale sequencing of environmental samples. Appl Environ Microbiol 71:5544–5550
Ravnskov S, Jakobsen I (1995) Functional compatibility in arbuscular mycorrhizas measured as hyphal P transport to the plant. New Phytol 129:611–618
Rossman AY, Tulloss RE, O’Dell TE, Thorn RG (1998) Protocols for an all taxa biodiversity inventory of fungi in a Costa Rican conservation area. Parkway, Boone
Ryberg M, Kristiansson E, Sjökvist E, Nilsson RH (2009) An outlook on the fungal internal transcribed spacer sequences in GenBank and the introduction of a web-based tool for the exploration of fungal diversity. New Phytol 181:471–474
Scheublin TR, Ridgway KP, Young PW, van der Heijden MGA (2004) Nonlegumes, legumes, and nodules harbor different arbuscular mycorrhizal fungal communities. Appl Environ Microbiol 70:6240–6246
Smith SE, Read DJ (2008) Mycorrhizal symbiosis, 3rd edn. Academic, London
Unterseher M, Jumpponen A, Öpik M, Tedersoo L, Moora M, Dormann CF, Schnittler M (2011) Species abundance distributions and richness estimations in fungal metagenomics – lessons learned from community ecology. Mol Ecol 20:275–285
Vandenkoornhuyse P, Baldauf SL, Leyval C, Straczek J, Young JPW (2002) Extensive fungal diversity in plant roots. Science 295:2051
Vandenkoornhuyse P, Maheé S, Ineson P, Staddon P, Ostle N, Cliquet JB (2007) Active root-associated microbes identified by rapid incorporation of plant-derived carbon into RNA. Proc Natl Acad Sci USA 104:16970–16975
Voisin AS, Salon C, Jeudy C, Warembourg R (2003) Root and nodule growth in Pisum sativum L. in relation to photosynthesis: Analysis using 13 C-labelling. Ann Bot 92:557–563
Vrålstad T, Myhre E, Schumacher T (2002) Molecular diversity and phylogenetic affinities of symbiotic root-associated ascomycetes of the Helotiales in burnt and metal polluted habitats. New Phytol 155:131–148
Whipps JM (2004) Prospects and limitations for mycorrhizas in biocontrol of root pathogens. Can J Bot 82:1198–1225
Acknowledgements
We thank Karsten Malmskov (Ardo A/S) and Steen Meier for help sampling soil in the field, Anne-Pia Larsen and Jacob I. Bañuelos Trejo for assistance with root staining, Henriette Nyskjold for technical assistance, Bernd Wollenweber for critical review on the manuscript and Kirsten Jensen for linguistic editing.
Author information
Authors and Affiliations
Corresponding author
Additional information
Responsible Editor: Erik J. Joner.
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
(DOCX 28 kb)
Rights and permissions
About this article
Cite this article
Yu, L., Nicolaisen, M., Larsen, J. et al. Succession of root-associated fungi in Pisum sativum during a plant growth cycle as examined by 454 pyrosequencing. Plant Soil 358, 225–233 (2012). https://doi.org/10.1007/s11104-012-1188-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11104-012-1188-5