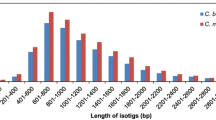
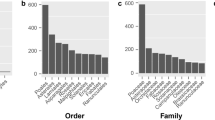
Abstract
Genes with similar or related functions in chloroplasts are often arranged in close proximity, forming clusters on chromosomes. These clusters are transcribed coordinated to facilitate the expression of genes with specific function. Our previous study revealed a significant negative correlation between the chloroplast gene expression level of the rare medicinal fern Ophioglossum vulgatum and its evolutionary rates as well as selection pressure. Therefore, in this study, we employed a combination of SMRT and Illumina sequencing technology to analyze the full-length transcriptome sequencing of O. vulgatum for the first time. In particular, we experimentally identified gene clusters based on transcriptome data and investigated the effects of chloroplast gene clustering on expression and evolutionary patterns. The results revealed that the total sequenced data volume of the full-length transcriptome of O. vulgatum amounted to 71,950,652,163 bp, and 110 chloroplast genes received transcript coverage. Nine different types of gene clusters were experimentally identified in their transcripts. The chloroplast cluster genes may cause a decrease in non-synonymous substitution rate and selection pressure, as well as a reduction in transversion rate, transition rate, and their ratio. While expression levels of chloroplast cluster genes in leaf, sporangium, and stem would be relatively elevated. The Mann–Whitney U test indicated statistically significant in the selection pressure, sporangia and leaves groups (P < 0.05). We have contributed novel full-length transcriptome data resources for ferns, presenting new evidence on the effects of chloroplast gene clustering on expression land evolutionary patterns, and offering new theoretical support for transgenic research through gene clustering.
Key message
The clustering of chloroplast genes in Ophioglossum vulgatum demonstrates a tendency towards elevated expression levels in sporangium, leaf, and stem, while exhibiting a decline in evolutionary rates and selection pressure.








Similar content being viewed by others
Data availability
The full-length transcriptome data were deposited in NCBI Sequence Read Archive (SRA) at (https://www.ncbi.nlm.nih.gov/bioproject), the Accession Number is PRJNA856114.
References
Adams MD, Kelley JM, Gocayne JD, Dubnick M, Polymeropoulos MH, Xiao H, Merril CR, Wu A, Olde B, Moreno RF, Kerlavage AR, McCombie WR, Venter JC (1991) Complementary DNA sequencing: expressed sequence tags and human genome project. Science 252:1651–1656. https://doi.org/10.1126/science.2047873
Anders S, McCarthy DJ, Chen Y, Okoniewski M, Smyth GK, Huber W, Robinson MD (2013) Count-based differential expression analysis of RNA sequencing data using R and bioconductor. Nat Protoc 8:1765–1786. https://doi.org/10.1038/nprot.2013.099
Boldogkői Z, Moldován N, Balázs Z, Snyder M, Tombácz D (2019) Long-read sequencing—a powerful tool in viral transcriptome research. Trends Microbiol 27:578–592. https://doi.org/10.1016/j.tim.2019.01.010
Börner T, Aleynikova AY, Zubo YO, Kusnetsov VV (2015) Chloroplast RNA polymerases: role in chloroplast biogenesis. Biochim Biophys Acta 1847:761–769. https://doi.org/10.1016/j.bbabio.2015.02.004
Branton D, Deamer DW, Marziali A, Bayley H, Benner SA, Butler T, Di Ventra M, Garaj S, Hibbs A, Huang X, Jovanovich SB, Krstic PS, Lindsay S, Ling XS, Mastrangelo CH, Meller A, Oliver JS, Pershin YV, Ramsey JM, Riehn R, Soni GV, Tabard-Cossa V, Wanunu M, Wiggin M, Schloss JA (2008) The potential and challenges of nanopore sequencing. Nat Biotechnol 26:1146–1153. https://doi.org/10.1038/nbt.1495
Byrne A, Cole C, Volden R, Vollmers C (2019) Realizing the potential of full-length transcriptome sequencing. Phil Trans R Soc B 374:20190097. https://doi.org/10.1098/rstb.2019.0097
Cai Z, Xie Z, Huang L, Wang Z, Pan M, Yu X, Xu S, Luo J (2022) Full-length transcriptome analysis of Adiantum flabellulatum gametophyte. PeerJ 10:e13079. https://doi.org/10.7717/peerj.13079
Cao Y, Xie L, Liu K, Liang Y, Dai X, Wang X, Lu J, Zhang X, Li X (2021) The antihypertensive potential of flavonoids from Chinese herbal medicine: a review. Pharmacol Res 174:105919. https://doi.org/10.1016/j.phrs.2021.105919
Cavaiuolo M, Kuras R, Wollman F, Choquet Y, Vallon O (2017) Small RNA profiling in Chlamydomonas: insights into chloroplast RNA metabolism. Nucleic Acids Res 45:10783–10799. https://doi.org/10.1093/nar/gkx668
Chen C, Chen H, Zhang Y, Thomas HR, Frank MH, He Y, Xia R (2020) TBtools: an integrative toolkit developed for interactive analyses of big biological data. Mol Plant 13:1194–1202. https://doi.org/10.1016/j.molp.2020.06.009
Clericuzio M, Tinello S, Burlando B, Ranzato E, Martinotti S, Cornara L, La Rocca A (2012) Flavonoid oligoglycosides from Ophioglossum vulgatum L. having wound healing properties. Planta Med 78:1639–1644. https://doi.org/10.1055/s-0032-1315149
Dixit R, Trivedi PK, Nath P, Sane PV (1999) Organization and post-transcriptional processing of the psb B operon from chloroplasts of Populus deltoides. Curr Genet 36:165–172. https://doi.org/10.1007/s002940050487
Field KJ, Leake JR, Tille S, Allinson KE, Rimington WR, Bidartondo MI, Beerling DJ, Cameron DD (2015) From mycoheterotrophy to mutualism: mycorrhizal specificity and functioning in Ophioglossum vulgatum sporophytes. New Phytol 205:1492–1502. https://doi.org/10.1111/nph.13263
Halbeisen RE, Galgano A, Scherrer T, Gerber AP (2008) Post-transcriptional gene regulation: from genome-wide studies to principles. Cell Mol Life Sci 65:798–813. https://doi.org/10.1007/s00018-007-7447-6
Hao J, Liang Y, Zhu M, Ping J, Feng P, Su Y, Wang T (2021) The complete chloroplast genome of Ophioglossum vulgatum L. (Ophioglossaceae) and phylogenetic analysis. Mitochondrial DNA B 6:2730–2731. https://doi.org/10.1080/23802359.2021.1966333
Hao J, Liang Y, Ping J, Li J, Shi W, Su Y, Wang T (2022a) Chloroplast gene expression level is negatively correlated with evolutionary rates and selective pressure while positively with codon usage bias in Ophioglossum vulgatum L. BMC Plant Biol 22:580. https://doi.org/10.1186/s12870-022-03960-8
Hao J, Liang Y, Su Y, Wang T (2022b) The complete mitochondrial genome of Ophioglossum vulgatum L. is with highly repetitive sequences: intergenomic fragment transfer and phylogenetic analysis. Genes 13:1287. https://doi.org/10.3390/genes13071287
He L, Fu S, Xu Z, Yan J, Xu J, Zhou H, Zhou J, Chen X, Li Y, Au KF, Yao H (2017) Hybrid sequencing of full-length cDNA transcripts of stems and leaves in Dendrobium officinale. Genes 8:257. https://doi.org/10.3390/genes8100257
He Z, Su Y, Wang T (2021) Full-length transcriptome analysis of four different tissues of Cephalotaxus oliveri. Int J Mol Sci 22:787. https://doi.org/10.3390/ijms22020787
Hihara Y, Kamei A, Kanehisa M, Kaplan A, Ikeuchi M (2001) DNA microarray analysis of cyanobacterial gene expression during acclimation to high light. Plant Cell 13:793–806. https://doi.org/10.1105/tpc.13.4.793
Hong Y, Wang Z, Li M, Su Y, Wang T (2022) First multi-organ full-length transcriptome of tree fern Alsophila spinulosa highlights the stress-resistant and light-adapted genes. Front Genet 12:784546. https://doi.org/10.3389/fgene.2021.784546
Hu B, Jin J, Guo AY, Zhang H, Luo J, Gao G (2015) GSDS 2.0: an upgraded gene feature visualization server. Bioinformatics 31:1296–1297. https://doi.org/10.1093/bioinformatics/btu817
Hunt M, Silva ND, Otto TD, Parkhill J, Keane JA, Harris SR (2015) Circlator: automated circularization of genome assemblies using long sequencing reads. Genome Biol 16:294. https://doi.org/10.1186/s13059-015-0849-0
Jansen RK, Ruhlman TA (2012) Plastid genomes of seed plants. In: Bock R, Knoop V (eds) Genomics of chloroplasts and mitochondria, advances in photosynthesis and respiration 35. Springer, Dordrecht, pp 103–126. https://doi.org/10.1007/978-94-007-2920-9_5
Kang YJ, Yang DC, Kong L, Hou M, Meng YQ, Wei L, Gao G (2017) CPC2: a fast and accurate coding potential calculator based on sequence intrinsic features. Nucleic Acids Res 45:W12–W16. https://doi.org/10.1093/nar/gkx428
Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, Thierer T, Ashton B, Meintjes P, Drummond A (2012) Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28:1647–1649. https://doi.org/10.1093/bioinformatics/bts199
Khandelwal S (1990) Chromosome evolution in the genus Ophioglossum L. Bot J Linn Soc 102:205–217
Kim D, Langmead B, Salzberg SL (2015) HISAT: a fast spliced aligner with low memory requirements. Nat Methods 12:357–360. https://doi.org/10.1038/nmeth.3317
Kumar S, Stecher G, Tamura K (2016) MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33:1870–1874. https://doi.org/10.1093/molbev/msw054
Langmead B, Salzberg SL (2012) Fast gapped-read alignment with Bowtie 2. Nat Methods 9:357–359. https://doi.org/10.1038/nmeth.1923
Legen J, Ruf S, Kroop X, Wang G, Barkan A, Bock R, Schmitz-Linneweber C (2018) Stabilization and translation of synthetic operon-derived mRNAs in chloroplasts by sequences representing PPR protein-binding sites. Plant J 94:8–21. https://doi.org/10.1111/tpj.13863
Letunic I, Bork P (2021) Interactive Tree Of Life (iTOL) v5: an online tool for phylogenetic tree display and annotation. Nucleic Acids Res 49:W293–W296. https://doi.org/10.1093/nar/gkab301
Li B, Dewey CN (2011) RSEM: accurate transcript quantification from RNA-seq data with or without a reference genome. BMC Bioinform 12:323. https://doi.org/10.1186/1471-2105-12-323
Li FW, Kuo LY, Rothfels CJ, Ebihara A, Chiou WL, Windham MD, Pryer KM (2011) rbcL and matK earn two thumbs up as the core DNA barcode for ferns. PLoS ONE 6:e26597. https://doi.org/10.1371/journal.pone.0026597
Li Y, Wang S, Aioub AAA, Qie X, Wu W, Hu Z (2019) Identification and analysis of full-length transcripts involved in the biosynthesis of insecticidal lignan (+)-haedoxan A in Phryma leptostachya. Ind Crops Prod 142:111868. https://doi.org/10.1016/j.indcrop.2019.111868
Macedo-Osorio KS, Pérez-España VH, Garibay-Orijel C, Guzmán-Zapata D, Durán-Figueroa NV, Badillo-Corona JA (2018) Intercistronic expression elements (IEE) from the chloroplast of Chlamydomonas reinhardtii can be used for the expression of foreign genes in synthetic operons. Plant Mol Biol 98:303–317. https://doi.org/10.1007/s11103-018-0776-z
McCarthy A (2010) Third generation DNA sequencing: Pacific biosciences’ single molecule real time technology. Chem Biol 17:675–676. https://doi.org/10.1016/j.chembiol.2010.07.004
Nguyen LT, Schmidt HA, Von Haeseler A, Minh BQ (2015) IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol 32:268–274. https://doi.org/10.1093/molbev/msu300
Nützmann HW, Scazzocchio C, Osbourn A (2018) Metabolic gene clusters in eukaryotes. Annu Rev Genet 52:159–183. https://doi.org/10.1146/annurev-genet-120417-031237
Obata T, Kobayashi K, Tadakuma R, Akasaka T, Iba K, Negi J (2021) The endoplasmic reticulum pathway for membrane lipid synthesis has a significant contribution toward shoot removal-induced root chloroplast development in Arabidopsis. Plant Cell Physiol 62:494–501. https://doi.org/10.1093/pcp/pcab009
Pertea M, Pertea GM, Antonescu CM, Chang TC, Mendell JT, Salzberg SL (2015) StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat Biotechnol 33:290–295. https://doi.org/10.1038/nbt.3122
Ping J, Zhong X, Wang T, Su Y (2023) The broken chloroplast gene clusters in gymnosperms exhibit elevated substitution rates. Forests 14:1681. https://doi.org/10.3390/f14081681
Pond SLK, Frost SDW, Muse SV (2005) HyPhy: hypothesis testing using phylogenies. Bioinformatics 21:676–679. https://doi.org/10.1093/bioinformatics/bti079
PPG I (2016) A community-derived classification for extant lycophytes and ferns. J Syst Evol 54:563–603. https://doi.org/10.1111/jse.12229
Razin SV, Ioudinkova ES, Kantidze OL, Iarovaia OV (2021) Co-regulated genes and gene clusters. Genes 12:907. https://doi.org/10.3390/genes12060907
Robinson MD, Oshlack A (2010) A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol 11:R25. https://doi.org/10.1186/gb-2010-11-3-r25
Robinson MD, McCarthy DJ, Smyth GK (2010) edgeR: a bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26:139–140. https://doi.org/10.1093/bioinformatics/btp616
Salmela L, Rivals E (2014) LoRDEC: accurate and efficient long read error correction. Bioinformatics 30:3506–3514. https://doi.org/10.1093/bioinformatics/btu538
Sanger F, Nicklen S, Coulson AR (1977) DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA 74:5463–5467. https://doi.org/10.1073/pnas.74.12.5463
Shahar N, Weiner I, Stotsky L, Tuller T, Yacoby I (2019) Prediction and large-scale analysis of primary operons in plastids reveals unique genetic features in the evolution of chloroplasts. Nucleic Acids Res 47:3344–3352. https://doi.org/10.1093/nar/gkz151
Shinohara W, Nakato N, Yatabe-Kakugawa Y, Oka T, Kim JK, Murakami N, Noda H, Sahashi N (2013) The use of matK in Ophioglossaceae phylogeny and the determination of Mankyua chromosome number shed light on chromosome number evolution in Ophioglossaceae. Syst Bot 38:564–570. https://doi.org/10.1600/036364413X670232
Sun M, Li J, Li D, Huang F, Wang D, Li H, Xing Q, Zhu H, Shi L (2018) Full-length transcriptome sequencing and modular organization analysis of the naringin/neoeriocitrin-related gene expression pattern in Drynaria roosii. Plant Cell Physiol 59:1398–1414. https://doi.org/10.1093/pcp/pcy072
Tanaka M, Wakasugi T, Sugita M, Shinozaki K, Sugiura M (1986) Genes for the eight ribosomal proteins are clustered on the chloroplast genome of tobacco (Nicotiana tabacum): similarity to the S10 and spc operons of Escherichia coli. Proc Natl Acad Sci USA 83:6030–6034. https://doi.org/10.1073/pnas.83.16.6030
Trincado JL, Entizne JC, Hysenaj G, Singh B, Skalic M, Elliott DJ, Eyras E (2018) SUPPA2: fast, accurate, and uncertainty-aware differential splicing analysis across multiple conditions. Genome Biol 19:40. https://doi.org/10.1186/s13059-018-1417-1
Velculescu VE, Zhang L, Vogelstein B, Kinzler KW (1995) Serial analysis of gene expression. Science 270:484–487. https://doi.org/10.1126/science.270.5235.484
Velculescu VE, Zhang L, Zhou W, Vogelstein J, Basrai MA, Bassett DE, Hieter P, Vogelstein B, Kinzler KW (1997) Characterization of the yeast transcriptome. Cell 88:243–251. https://doi.org/10.1016/S0092-8674(00)81845-0
Warden CD, Yuan YC, Wu X (2013) Optimal calculation of RNA-seq fold-change values. Int J Comput Bioinform In Silico Model 2:285–292
Wicke S, Schneeweiss GM, dePamphilis CW, Müller KF, Quandt D (2011) The evolution of the plastid chromosome in land plants: gene content, gene order, gene function. Plant Mol Biol 76:273–297. https://doi.org/10.1007/s11103-011-9762-4
Ye J, Cheng S, Zhou X, Chen Z, Kim SU, Tan J, Zheng J, Xu F, Zhang W, Liao Y, Zhu Y (2019) A global survey of full-length transcriptome of Ginkgo biloba reveals transcript variants involved in flavonoid biosynthesis. Ind Crops Prod 139:111547. https://doi.org/10.1016/j.indcrop.2019.111547
Zhang D, Gao F, Jakovlić I, Zou H, Zhang J, Li WX, Wang GT (2020a) PhyloSuite: an integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Mol Ecol Resour 20:348–355. https://doi.org/10.1111/1755-0998.13096
Zhang Y, An D, Li C, Zhao Z, Wang W (2020b) The complete chloroplast genome of greater duckweed (Spirodela polyrhiza 7498) using PacBio long reads: insights into the chloroplast evolution and transcription regulation. BMC Genomics 21:76. https://doi.org/10.1186/s12864-020-6499-y
Zhang Y, Zhang A, Li X, Lu C (2020c) The role of chloroplast gene expression in plant responses to environmental stress. Int J Mol Sci 21:6082. https://doi.org/10.3390/ijms21176082
Zhao L, Zhang H, Kohnen MV, Prasad KVSK, Gu L, Reddy ASN (2019) Analysis of transcriptome and epitranscriptome in plants using PacBio iso-seq and nanopore-based direct RNA sequencing. Front Genet 10:253. https://doi.org/10.3389/fgene.2019.00253
Zhou DX, Quigley F, Massenet O, Mache R (1989) Cotranscription of the S10- and spc-like operons in spinach chloroplasts and identification of three of their gene products. Mol Gen Genet 216:439–445. https://doi.org/10.1007/BF00334388
Zupok A, Kozul D, Schöttler MA, Niehörster J, Garbsch F, Liere K, Fischer A, Zoschke R, Malinova I, Bock R, Greiner S (2021) A photosynthesis operon in the chloroplast genome drives speciation in evening primroses. Plant Cell 33:2583–2601. https://doi.org/10.1093/plcell/koab155
Acknowledgements
We appreciate the platform and equipment provided by our laboratory.
Funding
This work was supported by the National Natural Science Foundation of China (31872670 and 32071781), Guangdong Basic and Applied Basic Research Foundation (2021A1515010911), Science and Technology Projects in Guangzhou (202206010107), and Project of Department of Science and Technology of Shenzhen City, Guangdong, China (JCYJ20190813172001780 and JCYJ20210324141000001).
Author information
Authors and Affiliations
Contributions
TW and YS conceived and designed the research. JH and YL contributed to the genome sequencing. JH, YL, and JP analyzed the data. JH drew the maps and wrote the manuscript. TW and YS revised the manuscript. All authors approved the final version of the article.
Corresponding authors
Ethics declarations
Conflict of interest
The authors have no conflict of interest to declare.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Hao, J., Liang, Y., Ping, J. et al. Full-length transcriptome analysis of Ophioglossum vulgatum: effects of experimentally identified chloroplast gene clusters on expression and evolutionary patterns. Plant Mol Biol 114, 31 (2024). https://doi.org/10.1007/s11103-024-01423-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11103-024-01423-2