Abstract
Key message
Total of 14 SNPs associated with overwintering-related traits and 75 selective regions were detected. Important candidate genes were identified and a possible network of cold-stress responses in woody plants was proposed.
Abstract
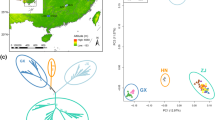
Local adaptation to low temperature is essential for woody plants to against changeable climate and safely survive the winter. To uncover the specific molecular mechanism of low temperature adaptation in woody plants, we sequenced 134 core individuals selected from 494 paper mulberry (Broussonetia papyrifera), which naturally distributed in different climate zones and latitudes. The population structure analysis, PCA analysis and neighbor-joining tree analysis indicated that the individuals were classified into three clusters, which showed forceful geographic distribution patterns because of the adaptation to local climate. Using two overwintering phenotypic data collected at high latitudes of 40°N and one bioclimatic variable, genome–phenotype and genome–environment associations, and genome-wide scans were performed. We detected 75 selective regions which possibly undergone temperature selection and identified 14 trait-associated SNPs that corresponded to 16 candidate genes (including LRR-RLK, PP2A, BCS1, etc.). Meanwhile, low temperature adaptation was also supported by other three trait-associated SNPs which exhibiting significant differences in overwintering traits between alleles within three geographic groups. To sum up, a possible network of cold signal perception and responses in woody plants were proposed, including important genes that have been confirmed in previous studies while others could be key potential candidates of woody plants. Overall, our results highlighted the specific and complex molecular mechanism of low temperature adaptation and overwintering of woody plants.






Similar content being viewed by others
Data availability
The RAD-seq sequences underlying this study have been deposited in NCBI database under BioProject code: PRJNA635706. The re-sequencing sequences underlying this study have been deposited in NCBI database under BioProject code: PRJNA635453. The phenotypic data and other relevant data supporting the findings of this work are contained within this paper and its Supplementary Files.
References
Alberto FJ, Aitken SN, Alía R, González-Martínez SC, Hänninen H, Kremer A, Lefèvre F, Lenormand T, Yeaman S, Whetten R, Savolainen O (2013) Potential for evolutionary responses to climate change-evidence from tree populations. Glob Chang Biol 19(6):1645–1661
Alexander DH, Novembre J, Lange K (2009) Fast model-based estimation of ancestry in unrelated individuals. Genome Res 19(9):1655–1664
Ali M, Plattner S, Radakovic Z, Wieczorek K, Elashry A, Grundler FM, Ammelburg M, Siddique S, Bohlmann H (2013) An Arabidopsis ATPase gene involved in nematode-induced syncytium development and abiotic stress responses. Plant J 74(5):852–866
Barrett JC, Fry B, Maller J, Daly MJ (2005) Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics 21(2):263–265
Benkova E (2016) Plant hormones in interactions with the environment. Plant Mol Biol 91:597
Bo K, Wang H, Pan Y, Behera T, Pandey S, Wen C, Wang Y, Simon PW, Li Y, Chen J, Weng Y (2016) SHORT HYPOCOTYL1 encodes a SMARCA3-like chromatin remodeling factor regulating elongation. Plant Physiol 172(2):1273–1292
Bonan GB (2008) Forests and climate change: forcings, feedbacks, and the climate benefits of forests. Science 320(5882):1444–1449
Chang CS, Liu HL, Moncada X, Seelenfreund A, Seelenfreund D, Chung KF (2015) A holistic picture of Austronesian migrations revealed by phylogeography of Pacific paper mulberry. Proc Natl Acad Sci U S A 112(44):13537–13542
Chen X, Poorey K, Carver MN, Muller U, Bekiranov S, Auble DT, Brow DA (2017) Transcriptomes of six mutants in the Sen1 pathway reveal combinatorial control of transcription termination across the Saccharomyces cerevisiae genome. PLOS Genet 13(6):e1006863
Cui TZ, Smith PM, Fox JL, Khalimonchuk O, Winge DR (2012) Late-stage maturation of the rieske Fe/S protein: Mzm1 stabilizes Rip1 but does not facilitate its translocation by the AAA ATPase Bcs1. Mol Cell Biol 32(21):4400–4409
Danecek P, Auton A, Abecasis G, Albers CA, Banks E, DePristo MA, Handsaker RE, Lunter G, Marth GT, Sherry ST, McVean G, Durbin R (2011) The variant call format and VCFtools. Bioinformatics 27(15):2156–2158
De La Torre AR, Wilhite B, Neale DB (2019) Environmental genome-wide association reveals climate adaptation is shaped by subtle to moderate allele frequency shifts in loblolly pine. Genome Biol Evol 11(10):2976–2989
Díaz-Arce N, Arrizabalaga H, Murua H, Irigoien X, Rodríguez-Ezpeleta N (2016) RAD-seq derived genome-wide nuclear markers resolve the phylogeny of tunas. Mol Phylogenet Evol 102:202–207
Ding Y, Shi Y, Yang S (2020) Molecular regulation of plant responses to environmental temperatures. Mol Plant 13:544–564
Du Z, Zhou X, Ling Y, Zhang Z, Su Z (2010) agriGO: a GO analysis toolkit for the agricultural community. Nucleic Acids Res 38(Web Server issue): W64-W70
Evans LM, Slavov GT, Rodgers-Melnick E, Martin J, Ranjan P, Muchero W, Brunner AM, Schackwitz W, Gunter L, Chen JG, Tuskan GA, DiFazio SP (2014) Population genomics of Populus trichocarpa identifies signatures of selection and adaptive trait associations. Nat Genet 46(10):1089–1096
Fan ZQ, Chen JY, Kuang JF, Lu WJ, Shan W (2017) The banana fruit SINA ubiquitin ligase MaSINA1 regulates the stability of MaICE1 to be negatively involved in cold stress response. Front Plant Sci 8:995
Felsenstein J (1989) PHYLIP: phylogeny inference package (version 3.2). Cladistics 5:164–166
Gao F, Ma P, Wu Y, Zhou Y, Zhang G (2019) Quantitative proteomic analysis of the response to cold stress in jojoba, a tropical woody crop. Int J Mol Sci 20(2):243
Harholt J, Jensen JK, Verhertbruggen Y, Søgaard C, Bernard S, Nafisi M, Poulsen CP, Geshi N, Sakuragi Y, Driouich A, Knox JP, Scheller HV (2012) ARAD proteins associated with pectic Arabinan biosynthesis form complexes when transiently overexpressed in planta. Planta 236(1):115–128
Hu YM, Peng XJ, Shen SH (2018) Multiculture and values of the paper mulberry from the perspective of ancient China’s science and technology. J Beijing For Univ 17(2):38–43
Huang J, Zhang J, Li W, Hu W, Duan L, Feng Y, Qiu F, Yue B (2013) Genome-wide association analysis of ten chilling tolerance indices at the germination and seedling stages in maize. J Integr Plant Biol 55(8):735–744
Ivanov A, Shuvalova E, Egorova T, Shuvalov A, Sokolova E, Bizyaev N, Shatsky I, Terenin I, Alkalaeva E (2019) Polyadenylate-binding protein-interacting proteins PAIP1 and PAIP2 affect translation termination. J Biol Chem 294(21):8630–8639
Kalberer SR, Leyva-Estrada N, Krebs SL, Arora R (2007) Frost dehardening and rehardening of floral buds of deciduous azaleas are influenced by genotypic biogeography. Environ Exp Bot 59(3):264–275
Kim BH, Kim SY, Nam KH (2012) Genes encoding plant-specific class III peroxidases are responsible for increased cold tolerance of the brassinosteroid-insensitive 1 mutant. Mol Cells 34(6):539
Kumar S, Stecher G, Tamura K (2016) MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33(7):1870–1874
Lasky JR, Upadhyaya HD, Ramu P, Deshpande S, Hash CT, Bonnette J, Juenger TE, Hyma K, Acharya C, Mitchell SE (2015) Genome–environment associations in sorghum landraces predict adaptive traits. Sci Adv 1(6):e1400218
Li H, Durbin R (2009) Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 25(14):1754–1760
Li P, Li YJ, Zhang FJ, Zhang GZ, Jiang XY, Yu HM, Hou BK (2017) The Arabidopsis UDP-glycosyltransferases UGT79B2 and UGT79B3, contribute to cold, salt and drought stress tolerance via modulating anthocyanin accumulation. Plant J 89:85–103
Li J, Chen GB, Rasheed A, Li D, Sonder K, Espinosa CZ, Wang J, Costich DE, Schnable PS, Hearne SJ, Li H (2019a) Identifying loci with breeding potential across temperate and tropical adaptation via EigenGWAS and EnvGWAS. Mol Ecol 28(15):3544–3560
Li Y, Cao K, Zhu G, Fang W, Chen C, Wang X, Zhao P, Guo J, Ding T, Guan L, Zhang Q, Guo W, Fei Z, Wang L (2019b) Genomic analyses of an extensive collection of wild and cultivated accessions provide new insights into peach breeding history. Genome Biol 20(1):36
Li Y, Wang Y, Tan S, Li Z, Yuan Z, Glanc M, Domjan D, Wang K, Xuan W, Guo Y, Gong Z, Friml J, Zhang J (2020) Root growth adaptation is mediated by PYLs ABA receptor-PP2A protein phosphatase complex. Adv Sci 7(3):1901455
Liang ZC, Duan SC, Sheng J, Zhu SS, Ni XM, Shao JH, Liu CH, Nick P, Du F, Fan PG, Mao RZ, Zhu YF, Deng WP, Yang M, Huang HC, Liu YX, Ding YQ, Liu XJ, Jiang JF, Zhu YY, Li SH, He XH, Chen W, Dong Y (2019) Whole-genome resequencing of 472 Vitis accessions for grapevine diversity and demographic history analyses. Nat Commun 10:1190
Lin F, Li S, Wang K, Tian H, Gao J, Zhao Q, Du C (2020) A Leucine-rich repeat receptor-like kinase, OsSTLK, modulates salt tolerance in rice. Plant Sci 296:110465
Liu X, Huang M, Fan B, Buckler ES, Zhang Z (2016) Iterative usage of fixed and random effect models for powerful and efficient genome-wide association studies. PLoS Genet 12(3):e1005767
Liu P, Du L, Huang Y, Gao S, Yu M (2017a) Origin and diversification of leucine-rich repeat receptor-like protein kinase (LRR-RLK) genes in plants. BMC Evol Biol 17(1):47
Liu Z, Jia Y, Ding Y, Shi Y, Li Z, Guo Y, Gong Z, Yang S (2017b) Plasma membrane CRPK1-mediated phosphorylation of 14-3-3 proteins induces their nuclear import to fine-tune CBF signaling during cold response. Mol Cell 66(1):117–128
Lobo A, Hansen OK, Hansen JK, Erichsen EO, Jacobsen B, Kjaer ED (2018) Local adaptation through genetic differentiation in highly fragmented Tilia cordata populations. Ecol Evol 8(12):5968–5976
Luo J, Shen G, Yan J, He C, Zhang H (2010) AtCHIP functions as an E3 ubiquitin ligase of protein phosphatase 2A subunits and alters plant response to abscisic acid treatment. Plant J 46(4):649–657
Ma Y, Dai X, Xu Y, Luo W, Zheng X, Zeng D, Pan Y, Lin X, Liu H, Zhang D (2015) COLD1 confers chilling tolerance in rice. Cell 160(6):1209–1221
Mathe C, Garda T, Freytag C, Mhamvas M (2019) The role of serine-threonine protein phosphatase PP2A in plant oxidative stress signaling-facts and hypotheses. Int J Mol Sci 20(12):3028
Mckenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, Depristo MA (2010) The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 20(9):1297–1303
Montwé D, Isaac-Renton M, Hamann A, Spiecker H (2018) Cold adaptation recorded in tree rings highlights risks associated with climate change and assisted migration. Nat Commun 9(1):1574
Moustafa K, AbuQamar S, Jarrar M, Al-Rajab AJ, Trémouillaux-Guiller J (2014) MAPK cascades and major abiotic stresses. Plant Cell Rep 33(8):1217–1225
Palm-Forster M, Eschen-Lippold L, Uhrig J, Scheel D, Lee J (2017) A novel family of proline/serine-rich proteins, which are phospho-targets of stress-related mitogen-activated protein kinases, differentially regulates growth and pathogen defense in Arabidopsis thaliana. Plant Mol Biol 95(1–2):123–140
Peñailillo J, Olivares G, Moncada X, Payacán C, Chang CS, Chung KF, Matthews PJ, Seelenfreund A, Seelenfreund D (2016) Sex distribution of paper mulberry (Broussonetia papyrifera) in the Pacific. PLoS ONE 11(8):e0161148
Peng XJ, Shen SH (2018) The paper mulberry: a novel model system for woody plant research. Chin Bull Bot 53(3):372–381
Peng XJ, Liu H, Chen PL, Tang F, Hu YM, Wang FF, Pi Z, Zhao ML, Chen NZ, Chen H, Zhang XK, Yan XQ, Liu M, Fu XJ, Zhao GF, Yao P, Wang LL, Dai H, Li XM, Xiong W, Xu WC, Zheng HK, Yu HY, Shen SH (2019) A chromosome-scale genome assembly of paper mulberry (Broussonetia papyrifera) provides new insights into its forage and papermaking usage. Mol Plant 12(2):661–667
Pi Z, Zhao ML, Peng XJ, Shen SH (2017) Phosphoproteomic analysis of paper mulberry reveals phosphorylation functions in chilling tolerance. J Proteome Res 16(5):1944–1961
Pluess AR, Frank A, Heiri C, Lalagüe H, Vendramin GG, Oddou-Muratorio S (2016) Genome-environment association study suggests local adaptation to climate at the regional scale in Fagus sylvatica. New Phytol 210(2):589–601
Rafalski A, Michele M (2004) Corn and humans: recombination and linkage disequilibrium in two genomes of similar size. Trends Genet 20(2):103–111
Sallam A, Arbaoui M, El-Esawi M, Abshire N, Martsch R (2016) Identification and verification of QTL associated with frost tolerance using linkage mapping and GWAS in winter faba bean. Front Plant Sci 7:1098
Shakiba E, Edwards JD, Jodari F, Duke SE, Baldo AM, Korniliev P, Mccouch SR, Eizenga GC (2017) Genetic architecture of cold tolerance in rice (Oryza sativa) determined through high resolution genome-wide analysis. PLoS ONE 12(3):e0172133
Shin SY, Kim MH, Kim YH, Park HM, Yoon HS (2013) Co-expression of monodehydroascorbate reductase and dehydroascorbate reductase from Brassica rapa effectively confers tolerance to freezing-induced oxidative stress. Mol Cells 36(4):304–315
Slavov GT, DiFazio SP, Martin J, Schackwitz W, Muchero W, Rodgers-Melnick E, Lipphardt MF, Pennacchio CP, Hellsten U, Pennacchio LA, Gunter LE, Ranjan P, Vining K, Pomraning KR, Wilhelm LJ, Pellegrini M, Mockler TC, Freitag M, Geraldes A, El-Kassaby YA, Mansfield SD, Cronk QC, Douglas CJ, Strauss SH, Rokhsar D, Tuskan GA (2012) Genome resequencing reveals multiscale geographic structure and extensive linkage disequilibrium in the forest tree Populus trichocarpa. New Phytol 196(3):713–725
Varshney RK, Saxena RK, Upadhyaya HD, Khan AW, Yu Y, Kim C, Rathore A, Kim D, Kim J, An S, Kumar V, Anuradha G, Yamini KN, Zhang W, Muniswamy S, Kim JS, Penmetsa RV, Wettberg E, Datta SK (2017) Whole-genome resequencing of 292 pigeon pea accessions identifies genomic regions associated with domestication and agronomic traits. Nat Genet 49(7):1082
Wang KJ, Li XH, Liu Y (2012) Fine-scale phylogenetic structure and major events in the history of the current wild soybean (Glycine soja) and taxonomic assignment of semi-wild type (Glycine gracilis Skvortz.) within the Chinese subgenus Soja. J Hered 103(1):13–27
Wang J, Li C, Yao X, Liu S, Zhang P, Chen K (2018) The Antarctic moss leucine-rich repeat receptor-like kinase (PnLRR-RLK2) functions in salinity and drought stress adaptation. Polar Biol 41(2):353–364
Wang J, Lu X, Yin Z, Wang D, Wang S, Mu M, Chen X, Guo L, Fan W, Chen C, Ye W (2019a) Isolation and characterization of GhLEA3 gene from upland cotton and its expression in response to low temperature stress. Cott Sci 31(2):89–100
Wang L, Yao L, Hao X, Li N, Wang Y, Ding C, Lei L, Qian W, Zeng J, Yang Y (2019b) Transcriptional and physiological analyses reveal the association of ROS metabolism with cold tolerance in tea plant. Environ Exp Bot 160:45–58
Wei X, Lu W, Mao L, Han X, Wei X, Zhao X, Xia M, Xu C (2020) ABF2 and MYB transcription factors regulate feruloyl transferase FHT involved in ABA-mediated wound suberization of kiwifruit. J Exp Bot 71(1):305–317
Wisniewski M, Nassuth A, Arora R (2018) Cold hardiness in trees: a mini-review. Front Plant Sci 9:1394
Xie H, Han Y, Li X, Dai W, Song X, Olsen KM, Qiang S (2019) Climate-dependent variation in cold tolerance of weedy rice and rice mediated by OsICE1 promoter methylation. Mol Ecol 29(1):121–137
Xu Z, Dong M, Peng X, Ku W, Zhao Y, Yang G (2019) New insight into the molecular basis of cadmium stress responses of wild paper mulberry plant by transcriptome analysis. Ecotoxicol Environ Saf 171:301–312
Yang H, Wang K (2015) Genomic variant annotation and prioritization with ANNOVAR and wANNOVAR. Nat Protoc 10(10):1556–1566
Yang J, Lee SH, Goddard ME, Visscher PM (2011) GCTA: a tool for genome-wide complex trait analysis. Am J Hum Genet 88(1):76–82
Yang L, Wu K, Gao P, Liu X, Li G, Wu Z (2014) GsLRPK, a novel cold-activated leucine-rich repeat receptor-like protein kinase from Glycine soja, is a positive regulator to cold stress tolerance. Plant Sci 215(3):19–28
Yang X, Wang R, Hu Q, Li S, Mao X, Jing H, Zhao J, Hu G, Fu J, Liu C (2019) DlICE1, a stress-responsive gene from Dimocarpus longan, enhances cold tolerance in transgenic Arabidopsis. Plant Physiol Biochem 142:490–499
Yeaman S, Hodgins KA, Lotterhos KE, Suren H, Nadeau S, Degner JC, Nurkowski KA, Smets P, Wang T, Gray LK, Liepe KJ, Hamann A, Holliday JA, Whitlock MC, Rieseberg LH, Aitken SN (2016) Convergent local adaptation to climate in distantly related conifers. Science 353(6306):1431–1433
Zhang Q, Zhang H, Sun L, Fan G, Ye M, Jiang L, Liu X, Ma K, Shi C, Bao F (2018) The genetic architecture of floral traits in the woody plant Prunus mume. Nat Commun 9(1):1702
Zhang M, Suren H, Holliday JA (2019) Phenotypic and genomic local adaptation across latitude and altitude in Populus trichocarpa. Genome Biol Evol 11(8):2256–2272
Zhao X, Zheng F, Li Y, Hao J, Tang Z, Tian C, Yang Q, Zhu T, Diao C, Zhang C, Chen M, Hu S, Gao P, Zhang L, Liao Y, Yu W, Chen M, Zou L, Gao W, Deng W (2019) BPTF promotes hepatocellular carcinoma growth by modulating hTERT signaling and cancer stem cell traits. Redox Biol 20:427–441
Zhou M, Xu M, Wu L, Shen C, Ma H, Lin J (2014) CbCBF from Capsella bursa-pastoris enhances cold tolerance and restrains growth in Nicotiana tabacumby antagonizing with gibberellin and affecting cell cycle signaling. Plant Mol Biol 85(3):259–275
Zhou Z, Jiang Y, Wang Z, Gou Z, Lyu J, Li W, Yu Y, Shu L, Zhao Y, Ma Y, Fang C, Shen Y, Liu T, Li C, Li Q, Wu M, Wang M, Wu Y, Dong Y, Wan W, Wang X, Ding Z, Gao Y, Xiang H, Zhu B, Lee SH, Wang W, Tian Z (2015) Resequencing 302 wild and cultivated accessions identifies genes related to domestication and improvement in soybean. Nat Biotechnol 33(4):408–414
Zhu J, Zhang KX, Wang WS, Gong W, Liu WX, Chen HG, Xu HH, Lu YT (2015) Low temperature inhibits root growth by reducing auxin accumulation via ARR1/12. Plant Cell Physiol 56(4):727–736
Acknowledgements
We thank Hui Chen, Zhi Pi, Feng Tang, Xiaokang Zhang, Jie Hu, Jinshan Wang, Songwei Li and Pu Shu for their help of sample collection and cultivation in this study. This work was supported by the National Natural Science Foundation of China (31870247, 32070358, 31770360) and the Beijing Natural Science Foundation (6202026).
Author information
Authors and Affiliations
Contributions
Y.H. and X.P. performed the research, collected genetic and phenotypic data, analyzed the data, and drafted the manuscript. F.W., M.Z. and P.C. contributed to the acquisition of the climate and common garden data. S.S. conceived and designed the experiments. S.S. and X.P. provided funding. All authors approved the final manuscript.
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Hu, Y., Peng, X., Wang, F. et al. Natural population re-sequencing detects the genetic basis of local adaptation to low temperature in a woody plant. Plant Mol Biol 105, 585–599 (2021). https://doi.org/10.1007/s11103-020-01111-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11103-020-01111-x