Abstract
Key message
Mapping QTL for stem-related traits using RIL population with ultra-high density bin map can better dissect pleiotropic QTL controlling stem architecture that can provide valuable information for maize genetic improvement.
Abstract
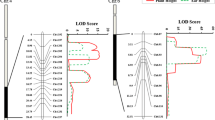
The maize stem is one of the most important parts of the plant and is also a component of many agronomic traits in maize. This study aimed to advance our understanding of the genetic mechanisms underlying maize stem traits. A recombinant inbred line (RIL) population derived from a cross between Ye478 and Qi319 was used to identify quantitative trait loci (QTL) controlling stem height (SH), ear height (EH), stem node number (SN), ear node (EN), and stem diameter (SD), and two derived traits (ear height coefficient (EHc) and ear node coefficient (ENc)). Using an available ultra-high density bin map, 46 putative QTL for these traits were detected on chromosomes 1, 3, 4, 5, 6, 7, 8, and 10. Individual QTL explained 3.5–17.7% of the phenotypic variance in different environments. Two QTL for SH, three for EH, two for EHc, one for SN, one for EN, and one for SD were detected in more than one environment. QTL with pleiotropic effects or multiple linked QTL were also identified on chromosomes 1, 3, 4, 6, 8, and 10, which are potential target regions for fine-mapping and marker-assisted selection in maize breeding programs. Further, we discussed segregation of bin markers (mk1630 and mk4452) associated with EHc QTL in the RIL population. We had identified two putative WRKY DNA-binding domain proteins, AC209050.3_FG003 and GRMZM5G851490, and a putative auxin response factor, GRMZM2G437460, which might be involved in regulating plant growth and development, as candidate genes for the control of stem architecture.



Similar content being viewed by others
Data availability
The raw sequence reads of RILs derived from Ye478 and Qi319 had been public on NCBI (https://www.ncbi.nlm.nih.gov/bioproject/627044, Accession: PRJNA627044).
Abbreviations
- SH:
-
Stem height
- EH:
-
Ear height
- EHc:
-
Ear height coefficient, EH/SH
- SN:
-
Stem node number
- EN:
-
Ear node
- ENc:
-
Ear node coefficient, EN/SN
- SD:
-
Stem diameter
- MAS:
-
Marker-assisted selection
- QTL:
-
Quantitative trait loci
- RIL:
-
Recombinant inbred line
- CTK:
-
Cytokinin
- IAA:
-
Aauxin
- GA:
-
Gibberellin
- BR:
-
Brassinosteroids
References
Beavis WD, Smith OS, Grant D, Fincher R (1994) Identification of quantitative trait loci using a small sample of topcrossed and F4 progeny from maize. Crop Sci 34:882–896
Bensen RJ, Johal GS, Crane VC, Tossberg JT, Schnable PS, Meeley RB, Briggs SP (1995) Cloning and characterization of the maize An1 gene. Plant Cell 7:75–84
Berke T, Rocheford T (1995) Quantitative trait loci for flowering, plant and ear height, and kernel traits in maize. Crop Sci 35(6):1542–1549
Bokore FE, Cuthbert RD, Knox RE, Singh A, Campbell HL, Pozniak CJ, N'Diaye A, Sharpe AG, Ruan Y (2019) Mapping quantitative trait loci associated with common bunt resistance in a spring wheat (Triticum aestivum L.) variety Lillian. Theor Appl Genet 132:3023–3033
Cai Y, Chen X, Xie K, Xing Q, Wu Y, Li J, Du C, Sun Z, Sun Z (2014) Dlf1, a WRKY transcription factor, is involved in the control of flowering time and plant height in rice. PLoS ONE 9:e102529
Chen Z, Wang B, Dong X, Liu H, Ren L, Chen J, Hauck A, Song W, Lai J (2014) An ultra-high density bin-map for rapid QTL mapping for tassel and ear architecture in a large F2 maize population. BMC Genomics 15:433
Cheng WS, Liu F, Li M, Hu XD, Chen H, Pappoe F (2015) Variation detection based on next-generation sequencing of type Chinese 1 strains of Toxoplasma gondii with different virulence from China. BMC Genomics 16:888
Collard B, Jahufer M, Brouwer J, Pang E (2005) An introduction to markers, quantitative trait loci (QTL) mapping and marker-assisted selection for crop improvement: The basic concepts. Euphytica 142:169–196
Feng G, Huang C, Xing J (2008) The research progress in lodging resistance of maize. Crops 4:12–14
Feng G, Jing X, Li Y, Wang L, Huang C (2010) Correlation and path analysis of lodging resistance with maize stem characters. Acta Agric Boreali-Sin 25:72–74
Fu Z, Shao K, Chen D, Wang B, Xu Z, Ding D (2011) Correlation analysis of the internode number above ear and lodging resistance in maize. J Henan Agric Univ 145:149–154
Fujioka S, Yamane H, Spray CR, Gaskin P, Macmillan J, Phinney BO, Takahashi N (1988) Qualitative and quantitative analyses of gibberellins in vegetative shoots of normal, dwarf-1, dwarf-2, dwarf-3, and dwarf-5 seedlings of Zea mays L. Plant Physiol 88:1367–1372
Guo D, Wang X, Han X, Wei B, Wang J, Li B, Yu H, Huang Q, Gu H, Qu L, Qin G (2015) The WRKY transcription factor WRKY71/EXB1 controls shoot branching by transcriptionally regulating RAX genes in Arabidopsis. Plant Cell 27:3112–3127
Hackenberg D, Asare-Bediako E, Baker A, Walley P, Jenner C, Greer S, Bramham L, Batley A, Edwards D, Delourme R, Barker G, Teakle G, Walsh J (2019) Identifcation and QTL mapping of resistance to Turnip yellows virus (TuYV) in oilseed rape, Brassica napus. Theor Appl Genet 133:383–393
Hittalmani S, Shashidhar H, Bagali P, Huang N, Sidhu J, Singh V, Khush G (2002) Molecular mapping of quantitative trait loci for plant growth, yield and yield related traits across three diverse locations in a doubled haploid rice population. Euphytica 125:207–214
Huang X, Ding F, Peng H-X, Pan J, He X, Xu J, Li L (2019) Research progress on family of plant WRKY transcription factors. Biotechnol Bull 35(12):129–143
Koester RP, Sisco PH, Stuber CW (1993) Identification of quantitative trait loci controlling days to flowering and plant height in two near isogenic lines of maize. Crop Sci 33:1209–1216
Li A, Dong H, Li H (2007) The study on combining ability of 6 exotic maize populations based on different heterotic backgrounds. J Hebei Agric Univ 30:13–18
Li D, Mao L, Yang J, Liu J, Zhang Y (2005) Breeding process and utilization of excellent maize inbred line 478. J Laiyang Agric Coll 22:159–164
Li Z, Zhang H, Wu X, Sun Y, Liu X (2014) Quantitative trait locus analysis for ear height in maize based on a recombinant inbred line population. Gene Mol Res 13:450–456
Lima M, Souza C, Bento D, Souza A, Garcia L (2006) Mapping QTL for grain yield and plant traits in a tropical maize population. Mol Breed 17:227–239
Liu K, Xu H, Liu G, Guan P, Zhou X, Peng H, Yao Y, Ni Z, Sun Q, Du J (2018) QTL mapping of flag leaf-related traits in wheat (Triticum aestivum L.). Theor Appl Genet 131:839–849
Liu T, Zhang J, Wang M, Wang Z, Li G, Qu L, Wang G (2007) Expression and functional analysis of ZmDWF4, an ortholog of Arabidopsis DWF4 from maize (Zea mays L.). Plant Cell Rep 26:2091–2099
Ma Y, Wang Q (2012) Research progress on the relationship between corn stalk traits and lodging resistance. Crops 2:10–15
Ma Y, Xue H, Zhang L, Zhang F, Ou C, Wang F, Zhang Z (2016) Involvement of auxin and brassinosteroid in dwarfism of autotetraploid apple (Malus × domestica). Sci Rep 6:26719
Melchinger AE, Utz HF, Schön CC (1998) Quantitative trait locus (QTL) mapping using different testers and independent population samples in maize reveals low power of QTL detection and large bias in estimates of QTL effects. Genetics 149:383–403
Michalczuk L (2002) Indole-3-acetic acid level in wood, bark and cambial sap of apple rootstocks differing in growth vigour. Acta Physiol Plant 24:131–136
Ribaut J, Betrán J (1999) Single large-scale marker-assisted selection (SLS-MAS). Mol Breed 5:531–541
Sakamoto T (2006) Phytohormones and rice crop yield: strategies and opportunities for genetic improvement. Transgenic Res 15:399–404
Sakamoto T, Miura K, Itoh H, Tatsumi T, Ueguchi-Tanaka M, Ishiyama K, Kobayashi M, Agrawal GK, Takeda S, Abe K, Miyao A, Hirochika H, Kitano H, Ashikari M, Matsuoka M (2004) An overview of gibberellin metabolism enzyme genes and their related mutants in rice. Plant Physiol 134:1642–1653
SAS Institute Inc (1999) SAS/STAT user’s guide, version 8. SAS Institute Inc, Cary, NC
Shang Q, Zhang D, Wang K, Wang G, Pan J, Li X, Shi L (2020) Diversity analysis of stalk traits in maize inbred lines in China. J Plant Genet Res 21:321–329
Soumelidou K, Morris DA, Battey NH, Barnett JR, John P (1994) Auxin transport capacity in relation to the dwarfing effect of apple rootstocks. J Horticult Sci 69:719–725
Spray CR, Kobayashi M, Suzuki Y (1996) The dwarf-1 (d1) mutant of Zea mays blocks three steps in the gibberellin-biosynthetic pathway. P Natl Acad Sci USA 93:10515–10518
Tanksley S (1993) Mapping polygenes. Annu Rev Genet 27:205–233
Teng F, Zhai L, Liu R, Bai W, Wang L, Huo D, Tao Y, Zheng Y, Zhang Z (2013) ZmGA3ox2, a candidate gene for a major QTL, qPH3.1, for plant height in maize. Plant J 73:405–416
van Ooijen J (2004) MapQTL® 5: software for the mapping of quantitative trait loci in experimental populations. Kyazma BV, Wageningen
Veldboom LR, Lee M (1994) Molecular-marker-facilitated studies of morphological traits in maize. 2: determination of QTLs for grain yield and yield components. Theor Appl Genet 894:451–458
Veldboom LR, Lee M (1996a) Genetic mapping of quantitative trait loci in maize in stress and nonstress environments. 1. Grain yield and yield components. Crop Sci 36:1310–1319
Veldboom LR, Lee M (1996b) Genetic mapping of quantitative trait loci in maize in stress and nonstress environments. 2. Plant Height Flower Crop Sci 36:1320–1327
Veldboom LR, Lee M, Woodman WL (1994) Molecular marker-facilitated studies in an elite maize population: 1. Linkage analysis and determination of QTL for morphological traits. Theor Appl Genet 88:7–16
VSN International Ltd (2008) GENSTAT for windows, 11th edn. VSN International, Hemel Hempstead
Wang T, Wang M, Hu S, Xiao Y, Tong H, Pan Q, Xue J, Yan J, Li J, Yang X (2015) Genetic basis of maize kernel starch content revealed by high-density single nucleotide polymorphism markers in a recombinant inbred line population. BMC Plant Biol 288:1–12
Wei L, Zhang X, Zhang Z, Liu H, Lin Z (2018) A new allele of the Brachytic2 gene in maize can efficiently modify plant architecture. Heredity 121:75–86
Wu J, Liu C, Shi Y, Song Y, Zhang G, Ma Z, Wang T, Li Y (2005) QTL analysis of plant height and ear height in maize under different water regimes. J Plant Genet Res 6:266–271
Xing A, Gao Y, Ye L, Zhang W, Cai L, Ching A, Llaca V, Johnson B, Liu L, Yang X, Kang D, Yan J, Li J (2015) A rare SNP mutation in Brachytic2 moderately reduces plant height and increases yield potential in maize. J Exp Bot 66:3791–3802
Xu T, Zhou F, Huang L, Xie P, Guan Y, Lan C, Liu P (2019) Research advances in maize lodging resistance. Anhui Agric Sci Bull 25:29–32
Ye J (2000) The first inbred lines Qi319 immune to maize southern rust bred in China. Agric Sci China 33:110
Zhang Y (2010) Mapping of quantitative trait loci (QTL) and genetic basis for plant height and ear height in maize. Dissertation, Chinese Academy of Agricultural Sciences
Zhao B, Li J (2012) Regulation of brassinosteroid biosynthesis and inactivation. J Integr Plant Biol 54:746–759
Zhao J, Wang R (2009) Factors promoting the steady increase of American maize production and their enlightenments for China. J Maize Sci 17(156–159):163
Zheng L (2016) Construction of chromosome segment substitution line and QTL mapping of plant height in maize. Dissertation, Northeast Agricultural University
Zheng X, Zhao Y, Shan D, Shi K, Wang L, Li Q, Wang N, Zhou J, Yao J, Xue Y, Fang S, Chu J, Guo Y, Kong J (2018) MdWRKY9 overexpression confers intensive dwarfing in the M26 rootstock of apple by directly inhibiting brassinosteroid synthetase MdDWF4 expression. New Phytol 217:1086–1098
Zheng ZP, Liu XH (2013) Genetic analysis of agronomic traits associated with plant architecture by QTL mapping in maize. Gene Mol Res 12:1243–1253
Zhou Z, Zhang C, Lu X, Wang L, Hao Z, Li M, Zhang D, Yong H, Weng J, Li X (2018) Dissecting the genetic basis underlying combining ability of plant height related traits in maize. Front Plant Sci 9:1117
Zhou Z, Zhang C, Zhou Y, Hao Z, Wang Z, Zeng X, Di H, Li M, Zhang D, Yong H, Zhang S, Weng J, Li X (2016) Genetic dissection of maize plant architecture with an ultra-high density bin map based on recombinant inbred lines. BMC Genomics 17:178
Acknowledgements
This research was jointly supported by the National Natural Science Foundation of China (31671706) and the Beijing Education Commission-General Project (KM201810020005).
Funding
This research was jointly supported by the National Natural Science Foundation of China (31671706) and the Beijing Education Commission-General Project (KM201810020005).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflicts of interest
The authors declare that they have no competing interests.
Ethical approval
I would like to declare on behalf of my co-authors that the work described is original, previously unpublished research. All the authors listed have approved the enclosed manuscript.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Shang, Q., Zhang, D., Li, R. et al. Mapping quantitative trait loci associated with stem-related traits in maize (Zea mays L.). Plant Mol Biol 104, 583–595 (2020). https://doi.org/10.1007/s11103-020-01062-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11103-020-01062-3