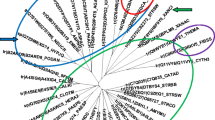
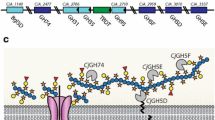
Abstract
Key message
The knowledge of substrate specificity of XET enzymes is important for the general understanding of metabolic pathways to challenge the established notion that these enzymes operate uniquely on cellulose-xyloglucan networks.
Abstract
Xyloglucan xyloglucosyl transferases (XETs) (EC 2.4.1.207) play a central role in loosening and re-arranging the cellulose-xyloglucan network, which is assumed to be the primary load-bearing structural component of plant cell walls. The sequence of mature TmXET6.3 from Tropaeolum majus (280 residues) was deduced by the nucleotide sequence analysis of complete cDNA by Rapid Amplification of cDNA Ends, based on tryptic and chymotryptic peptide sequences. Partly purified TmXET6.3, expressed in Pichia occurred in N-glycosylated and unglycosylated forms. The quantification of hetero-transglycosylation activities of TmXET6.3 revealed that (1,3;1,4)-, (1,6)- and (1,4)-β-d-glucooligosaccharides were the preferred acceptor substrates, while (1,4)-β-d-xylooligosaccharides, and arabinoxylo- and glucomanno-oligosaccharides were less preferred. The 3D model of TmXET6.3, and bioinformatics analyses of identified and putative plant xyloglucan endotransglycosylases (XETs)/hydrolases (XEHs) of the GH16 family revealed that H94, A104, Q108, K234 and K237 were the key residues that underpinned the acceptor substrate specificity of TmXET6.3. Compared to the wild-type enzyme, the single Q108R and K237T, and double-K234T/K237T and triple-H94Q/A104D/Q108R variants exhibited enhanced hetero-transglycosylation activities with xyloglucan and (1,4)-β-d-glucooligosaccharides, while those with (1,3;1,4)- and (1,6)-β-d-glucooligosaccharides were suppressed; the incorporation of xyloglucan to (1,4)-β-d-glucooligosaccharides by the H94Q variant was influenced most extensively. Structural and biochemical data of non-specific TmXET6.3 presented here extend the classic XET reaction mechanism by which these enzymes operate in plant cell walls. The evaluations of TmXET6.3 transglycosylation activities and the incidence of investigated residues in other members of the GH16 family suggest that a broad acceptor substrate specificity in plant XET enzymes could be more widespread than previously anticipated.









Similar content being viewed by others
Abbreviations
- 2GalManO6-OS:
-
2-galacto-manno-hexasaccharide
- Ara-OS6:
-
Arabino-heptasaccharide
- AraGal-OS:
-
Arabino-galacto-oligosaccharides
- AraXyl-OS:
-
Arabino-xylo-oligosaccharides
- Cello-OS:
-
Cello-oligosaccharides
- Cello-OS3:
-
Cello-triose
- Cello-OS4:
-
Cello-tetraose
- Cello-OS5:
-
Cello-pentaose
- Cello-OS6:
-
Cello-hexaose
- CMC:
-
Carboxymethyl cellulose
- cpk:
-
Atomic colour scheme
- C’XET:
-
TmXET6.3 without putative signal peptide
- GalUA-OS5:
-
Penta-galacturonic acid oligosaccharide
- GalMan-OS:
-
Galacto-manno-oligosaccharides
- GlcMan-OS:
-
Gluco-manno-oligosaccharides
- GH16:
-
Family 16 glycoside hydrolase
- HEC:
-
Hydroxyethyl cellulose
- HPLC:
-
High performance liquid chromatography
- La:
-
Laminarin
- La-OS:
-
Laminari-oligosaccharides
- Man-OS:
-
Manno-oligosaccharides
- Man-OS6:
-
Manno-hexa-oligosaccharide
- MLG-OS:
-
Mixed-linkage (1,3;1,4)-β-d-gluco-saccharides
- MLG-OS4 A, B, C:
-
(1,3;1,4)-β-d-tetra-glucosaccharides A, B, C
- MALDI:
-
Matrix-assisted laser desorption/ionisation
- MS:
-
Mass spectrometry
- OS:
-
Oligosaccharide(s)
- Pu:
-
Pustulan
- Pu-OS:
-
Pustulo-oligosaccharides
- TmXET6.3:
-
Tropaeolum majus XET6.3
- RACE:
-
Rapid Amplification of cDNA Ends
- RMSD:
-
Root-mean square deviation
- SDS-PAGE:
-
Sodium dodecyl sulfate–polyacrylamide gel electrophoresis
- SR:
-
Sulforhodamine
- TOF:
-
Time-of-flight
- XEH:
-
Xyloglucan endo-(1,4)-β-d-glucanase
- XET/XTH:
-
Xyloglucan endotransglycosylase/hydrolase
- XG:
-
Xyloglucan
- XG-OS7:
-
Xyloglucan heptasaccharide
- XG-OS8:
-
Xyloglucan octasaccharide
- XG-OS9:
-
Xyloglucan nonasaccharide
- XG-OS:
-
Xyloglucan oligosaccharides
- Xyl:
-
(1,4)-β-d-glucuronoxylan
- Xyl-OS:
-
(1,4)-β-d-glucurono-xylo-oligosaccharides
- WT:
-
Wild-type
References
Ait-Mohand F, Farkaš V (2006) Screening for hetero-transglycosylating activities in extracts from nasturtium (Tropaeolum majus). Carbohydr Res 341:577–581. https://doi.org/10.1016/j.carres.2006.01.018
Atkinson RG, Johnston SL, Yauk Y-K, Sharma NN, Schröder R (2009) Analysis of xyloglucan endotransglucosylase/hydrolase (XTH) gene families in kiwifruit and apple. Postharvest Biol Technol 51:149–157. https://doi.org/10.1016/j.postharvbio.2008.06.014
Baran B, Sulová Z, Stratilová E, Farkaš V (2000) Ping-pong character of nasturtium-seed xyloglucan endotransglycosylase (XET) reaction. Gen Phys Biophys 19:427–440
Baumann MJ, Eklöf JM, Michel G, Kallas ÅM, Teeri TT, Czjzek M, Brumer H (2007) Structural evidence for the evolution of xyloglucanase activity from xyloglucan endo-transglycosylases: biological implications for cell wall metabolism. Plant Cell 19:1947–1963. https://doi.org/10.1105/tpc.107.051391
Bencúrová M, Rendić D, Fabini G, Kopecky EM, Altman F, Wilson IBH (2003) Expression of eukaryotic glycosyltransferases in the yeast Pichia pastoris. Biochimie 85:413–422. https://doi.org/10.1016/S0300-9084(03)00072-5
Bourquin V, Nishikubo N, Abe H, Brumer H, Denman S, Eklund M, Christiernin M, Teeri TT, Sundberg B, Mellerowicz EJ (2002) Xyloglucan endotransglycosylases have a function during the formation of secondary cell walls of vascular tissues. Plant Cell 14:3073–3088. https://doi.org/10.1105/tpc.007773
Campbell P, Braam J (1999a) In vitro activities of four xyloglucan endotransglycosylases from Arabidopsis. Plant J 18:371–382. https://doi.org/10.1046/j.1365-313X.1999.00459.x
Campbell P, Braam J (1999b) Xyloglucan endotransglycosylases: diversity of genes, enzymes and potential wall-modifying functions. Trends Plant Sci 4:361–366. https://doi.org/10.1016/S1360-1385(99)01468-5
Carpita NC, Gibeaut DM (1993) Structural models of primary cell walls in flowering plants: consistency of molecular structure with the physical properties of the walls during growth. Plant J 3:1–30. https://doi.org/10.1111/j.1365-313X.1993.tb00007.x
Case DA, Babin V, Berryman JT, Betz RM, Cai Q, Cerutti DS, Cheatham TE, Darden TA, Duke RE, Gohlke H et al (2014) AMBER 14. University of California, San Francisco
Catalá C, Rose JKC, York WS, Albersheim P, Darvill AG, Bennett AB (2001) Characterization of a tomato xyloglucan endotransglycosylase gene that is down-regulated by auxin in etiolated hypocotyls. Plant Phys 127:1180–1192. https://doi.org/10.1104/pp.010481
Chanliaud E, Burrows KM, Jeronimidis G, Gidley MJ (2002) Mechanical properties of primary plant cell wall analogues. Planta 215:989–996. https://doi.org/10.1007/s00425-002-0783-8
Chanliaud E, De Silva J, Strongitharm B, Jeronimidis G, Gidley MJ (2004) Mechanical effects of plant cell wall enzymes on cellulose/xyloglucan composites. Plant J 38:27–37. https://doi.org/10.1111/j.1365-313X.2004.02018.x
Cosgrove DJ (2014) Re-constructing our models of cellulose and primary cell wall assembly. Curr Opin Plant Biol 22:122–131. https://doi.org/10.1016/j.pbi.2014.11.001
De Silva J, Jarman CD, Arrowsmith DA, Stronach MS, Chengappa S, Sidebottom C, Reid JSG (1993) Molecular characterization of a xyloglucan-specific endo-(1→4)-β-d-glucanase (xyloglucan endotransglycosylase) from nasturtium seeds. Plant J 3:701–711. https://doi.org/10.1046/j.1365-313X.1993.03050701.x
Fanutti C, Gidley MJ, Reid JSG (1993) Action of a pure xyloglucan endo-transglycosylase (formerly called xyloglucan-specific endo-(1–4)-β-d-glucanase) from the cotyledons of germinated nasturtium seeds. Plant J 3:691–700. https://doi.org/10.1111/j.1365-313X.1993.00691.x
Fanutti C, Gidley MJ, Reid JSG (1996) Substrate subsite recognition of the xyloglucan endo-transglycosylase or xyloglucan-specific endo-(1→)-β-d-glucanase from the cotyledons of germinated nasturtium (Tropaeolum majus L.) seeds. Planta 200:221–228. https://doi.org/10.1007/BF00208312
Farkaš V, Sulová Z, Stratilová E, Hanna R, Maclachlan G (1992) Cleavage of xyloglucan by nasturtium seed xyloglucanase and transglycosylation to xyloglucan subunit oligosaccharides. Arch Biochem Biophys 298:365–370. https://doi.org/10.1016/0003-9861(92)90423-T
Farrokhi N, Burton RA, Brownfield L, Hrmova M, Wilson SM, Bacic A, Fincher GB (2006) Plant cell wall biosynthesis: genetic, biochemical and functional genomics approaches to the identification of key genes. Plant Biotechnol J 4:145–167. https://doi.org/10.1111/j.1467-7652.2005.00169.x
Frohman MA, Dush MK, Martin GR (1998) Rapid production of full-lenght cDNAs from rare transcripts: amplification using a single gene-specific oligonucleotide primer. Proc Natl Acad Sci USA 85:8998–9002. https://doi.org/10.1073/pnas.85.23.8998
Fry SC (1997) Novel “dot-blot” assays for glycosyltransferases and glycosylhydrolases: optimization for xyloglucan endotransglycosylase (XET) activity. Plant J 11:1141–1150. https://doi.org/10.1046/j.1365-313X.1997.11051141.x
Fry SC, Smith RC, Renwick KF, Martin DJ, Hodge SK, Matthews KJ (1992) Xyloglucan endotransglycosylase, a new wall-loosening enzyme activity from plants. Biochem J 282:821–828. https://doi.org/10.1042/bj2820821
Fry SC, York WS, Albersheim P, Darvill A, Hayashi T, Joseleau J-P, Kato Y, Lorences EP, Maclachlan GA, McNeil M (1993) An unambiguous nomenclature for xyloglucan-derived oligosaccharides. Physiol Plant 89:1–3. https://doi.org/10.1111/j.1399-3054.1993.tb01778.x
Fry SC, Mohler KE, Nesselrode BHWA, Franková L (2008) Mixed-linkage β-glucan: xyloglucan endotransglucosylase, a novel wall-remodelling enzyme from Equisetum (horsetails) and charophytic algae. Plant J 55:240–252. https://doi.org/10.1111/j.1365-313X.2008.03504.x
Garajová S, Flodrová D, Ait-Mohand F, Farkaš V, Stratilová E (2008) Characterization of two partially purified xyloglucan endotransglycosylases from parsley (Petroselinum crispum) roots. Biologia 63:313–319. https://doi.org/10.2478/s11756-008-0067-2
Gupta R (2001) Prediction of glycosylation sites in proteomes: from post-translational modifications to protein function. Dissertation, DTU Bioinformatics, Denmark
Hayashi T (1989) Xyloglucans in the primary cell wall. Annu Rev Plant Physiol Plant Mol Biol 40:139–168. https://doi.org/10.1146/annurev.pp.40.060189.001035
Hrmova M, Farkaš V, Lahnstein J, Fincher GB (2007) A Barley xyloglucan xyloglucosyl transferase covalently links xyloglucan, cellulosic substrates, and (1,3;1,4)-β-d-glucans. J Biol Chem 282:12951–12962. https://doi.org/10.1074/jbc.M611487200
Hrmova M, Farkaš V, Harvey AJ, Lahnstein J, Wischmann B, Kaewthai N, Ezcurra I, Teeri TT, Fincher GB (2009) Substrate specificity and catalytic mechanism of a xyloglucan xyloglucosyl transferase HvXET6 from barley (Hordeum vulgare L.). FEBS J 276:437–456. https://doi.org/10.1111/j.1742-4658.2008.06791.x
Ibatullin FM, Banasiak A, Baumann MJ, Greffe L, Takahashi J, Mellerowicz EJ, Brumer H (2009) A Real-time fluorogenic assay for the visualization of glycoside hydrolase activity in planta. Plant Phys 151:1741–1750. https://doi.org/10.1104/pp.109.147439
Johansson P, Brumer H 3rd, Baumann MJ, Kallas AM, Henriksson H, Denman SE, Teeri TT, Jones TA (2004) Crystal structures of a poplar xyloglucan endotransglycosylase reveal details of transglycosylation acceptor binding. Plant Cell 16:874–886. https://doi.org/10.1105/tpc.020065
Johnston S, Prakash R, Chen NJ, Kumagai MH, Turano HM, Cooney JM, Atkinson RG, Paull RE, Cheetamun R, Bacic A et al (2013) An enzyme activity capable of endotransglycosylation of heteroxylan polysaccharides is present in plant primary cell walls. Planta 237:173–187. https://doi.org/10.1007/s00425-012-1766-z
Kim JS, Daniel G (2018) Heterogenous distribution of pectin and hemicellulose epitopes in the phloem of four hardwood species. Trees 32:393–414. https://doi.org/10.1007/s0046
Kosík O, Farkaš V (2008) One-pot synthesis of xyloglucan oligosaccharides fluorescently labeled with sulforhodamine. Anal Biochem 375:232–236. https://doi.org/10.1016/j.ab.2007.11.025
Kosík O, Auburn RP, Russel S, Stratilová E, Garajová S, Hrmova M, Farkaš V (2010) Polysaccharide microarrays for high-throughput screening of transglycosylase activities in plant extracts. Glycoconj J 27:79–87. https://doi.org/10.1007/s10719-009-9271-8
Kumar S, Stecher G, Tamura K (2016) MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol Biol Evol 33:1870–1874. https://doi.org/10.1093/molbev/msw054
Laemmli UK (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680–685. https://doi.org/10.1038/227680a0
Lu W, Wang Y, Jiang Y, Li J, Liu H, Duan X, Song L (2006) Differential expression of litchi XET genes in relation to fruit growth. Plant Phys Biochem 44:707–713. https://doi.org/10.1016/j.plaphy.2006.09.020
Mark P, Baumann MJ, Eklöf JM, Gullfot F, Michel G, Kallas ÅM, Teeri TT, Brumer H, Czjzek M (2009) Analysis of nastrutium TmNXG1 complexes by crystallography and molecular dynamics provides detailed insight into substrate recognition by family GH16 xyloglucan endo-transglycosylases and endo-hydrolases. Proteins 75:820–836. https://doi.org/10.1002/prot.22291
Mazáň M, Blanco N, Kováčová K, Firáková Z, Řehulka P, Farkaš V, Arroyo J (2013) A novel fluorescence assay and catalytic properties of Crh1 and Crh2 yeast cell wall transglycosylases. Biochem J 455:307–318. https://doi.org/10.1042/BJ20130354
McGregor N, Yin V, Tung ChCh, Van Petegem F, Brumer H (2017) Crystallographic insight into the evolutionary origins of xyloglucan endotransglycosylases and endohydrolases. Plant J 89:651–670. https://doi.org/10.1111/tpj.13421
Mohler KE, Simmons TJ, Fry SC (2013) Mixed-linkage glucan:xyloglucan endotransglucosylase (MXE) re-models hemicelluloses in Equisetum shoots but not in barley shoots or Equisetum callus. New Phytol 197:111–122. https://doi.org/10.1111/j.1469-8137.2012.04371.x
Muñoz-Bertomeu J, Miedes E, Lorences EP (2013) Expression of xyloglucan endotransglucosylase/hydrolase (XTH) genes and XET activity in ethylene treated apple and tomato fruits. J Plant Phys 170:1194–1201. https://doi.org/10.1016/j.jplph.2013.03.015
Nardi CF, Villarreal NM, Opazo MC, Martínez GA, Moya-León MA, Civello PM (2014) Expression of FaXTH1 and FaXTH2 genes in strawberry fruit. Cloning of promoter regions and effect of plant growth regulators. Sci Hortic 165:111–122. https://doi.org/10.1016/j.scienta.2013.10.035
Nei M, Kumar S (2000) Molecular evolution and phylogenetics. Oxford University Press, New York. https://doi.org/10.1046/j.1365-2540.2001.0923a.x
Nishikubo N, Awano T, Banasiak A, Bourquin V, Ibatullin F, Funada R, Brumer H, Teeri TT, Hayashi T, Sundberg B, Mellerowicz EJ (2007) Xyloglucan endo-transglycosylase (XET) functions in gelatinous layers of tension wood fibers in poplar—a glimpse into the mechanism of the balancing act of trees. Plant Cell Phys 48:843–855. https://doi.org/10.1093/pcp/pcm055
Nishikubo N, Takahashi J, Roos AA, Derba-Maceluch M, Piens K, Brumer H, Teeri TT, Stålbrand H, Mellerowicz EJ (2011) Xyloglucan endo-transglycosylase-mediated xyloglucan rearrangements in developing wood of hybrid aspen. Plant Phys 155:399–413. https://doi.org/10.1104/pp.110.166934
Nishitani K, Tominaga R (1992) Endo-xyloglucan transferase, a novel class of glycosyltransferase that catalyzes transfer of a segment of xyloglucan molecule to another xyloglucan molecule. J Biol Chem 267:21058–21064
Park YB, Cosgrove DJ (2015) Xyloglucan and its interactions with other components of the growing cell wall. Plant Cell Phys 56:180–194. https://doi.org/10.1093/pcp/pcu204
Peña MJ, Kong Y, York WS, O’Neill MA (2012) A galacturonic acid-containing xyloglucan is involved in Arabidopsis root hair tip growth. Plant Cell 24:4511–4524. https://doi.org/10.1105/tpc.112.103390
Petersen TN, Brunak S, von Heijne G, Nielsen H (2011) SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods 8:785–786. https://doi.org/10.1038/nmeth.1701
Raemaekers R, de Muro L, Gatehouse JA, Fordham-Skelton AP (1999) Functional phytohemagglutinin (PHA) and Galanthus nivalis agglutinin (GNA) expressed in Pichia pastoris. Eur J Biochem 265:394–403. https://doi.org/10.1046/j.1432-1327.1999.00749.x
Redgwell RJ, Fry SC (1993) Xyloglucan endotransglycosylase activity increases during kiwifruit (Actinidia deliciosa) ripening (implications for fruit softening). Plant Phys 103:1399–1406. https://doi.org/10.1104/pp.103.4.1399
Rose JKC, Bennet AB (1999) Cooperative disassembly of the cellulose–xyloglucan network of plant cell walls: parallels between cell expansion and fruit ripening. Trends Plant Sci 4:176–183. https://doi.org/10.1016/S1360-1385(99)01405-3
Rose TM, Schultz ER, Henikoff JG, Pietrokovski S, McCallum CM, Henikoff S (1998) Consensus-degenerate hybrid oligonucleotide primers for amplification of distantly related sequences. Nucleic Acids Res 26:1628–1635. https://doi.org/10.1093/nar/26.7.1628
Rose JKC, Braam J, Fry SC, Nishitani K (2002) The XTH family of enzymes involved in xyloglucan endotransglucosylation and endohydrolysis: current perspectives and a new unifying nomenclature. Plant Cell Phys 43:1421–1435. 10.1093%2Fpcp%2Fpcf171
Roth C, Moroz OV, Ariza A, Skov LK, Ayabe K, Davies GJ, Wilson KS (2018) Structural insight into industrially-relevant glucoamylases: flexible positions of starch-binding domains. Acta Crystallogr D 74:463–470. https://doi.org/10.1107/S2059798318004989
Ruprecht C, Dallabernardina P, Smith PJ, Urbanowicz BR, Pfrengle F (2018) Analyzing Xyloglucan endotransglycosylases by incorporating synthetic oligosaccharides into plant cell walls. ChemBioChem 19:793–798. https://doi.org/10.1002/cbic.201700638
Saitou N, Nei M (1987) The Neighbor-Joining method: A new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425. https://doi.org/10.1093/oxfordjournals.molbev.a040454
Šali A, Blundell TL (1993) Comparative protein modelling by satisfaction of spatial restraints. J Mol Biol 234:779–815. https://doi.org/10.1006/jmbi.1993.1626
Schröder R, Atkinson RG, Langenkämper G, Redgwell RJ (1998) Biochemical and molecular characterization of xyloglucan endotransglycosylase from ripe kiwifruit. Planta 204:242–251. https://doi.org/10.1007/s004250050253
Schröder R, Wegrzyn TF, Sharma NN, Atkinson RG (2006) LeMAN4 endo-beta-mannanase from ripe tomato fruit can act as a mannan transglycosylase or hydrolase. Planta 224:1091–1102. https://doi.org/10.1007/s00425-006-0286-0
Schröder R, Atkinson RG, Redgwell RJ (2009) Re-interpreting the role of endo-β-mannanases as mannan endotransglycosylase/hydrolases in the plant cell wall. Ann Bot 104:197–204. https://doi.org/10.1093/aob/mcp120
Shinohara N, Sunagawa N, Tamura S, Yokoyama R, Ueda M, Igarashi K, Nishitani K (2017) The plant cell-wall enzyme AtXTH3 catalyses covalent cross-linking between cellulose and cellooligosaccharide. Sci Rep 7:46099. https://doi.org/10.1038/srep46099
Sievers F, Wilm A, Dineen DG, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Söding J, Thompson JD, Higgins DG (2011) Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol 7:539. https://doi.org/10.1038/msb.2011.75
Simmons TJ, Fry SC (2017) Bonds broken and formed during the mixed-linkage glucan: xyloglucan endotransglucosylase reaction catalysed by Equisetum hetero-trans-β-glucanase. Biochem J 474:1055–1070. https://doi.org/10.1042/BCJ20160935
Simmons TJ, Mohler KE, Holland C, Goubet F, Franková L, Houston DR, Hudson AD, Meulewaeter F, Fry SC (2015) Hetero-trans-β-glucanase, an enzyme unique to Equisetum plants, functionalizes cellulose. Plant J 83:753–769. https://doi.org/10.1111/tpj.12935
Smith RC, Fry SC (1991) Endotransglycosylation of xyloglucans in plant cell suspension cultures. Biochem J 279:529–535. https://doi.org/10.1042/bj2790529
Steele NM, Sulová Z, Campbell P, Braam J, Farkaš V, Fry SC (2001) Ten isoenzymes of xyloglucan endotransglycosylase from plant cell walls select and cleave the donor substrate stochastically. Biochem J 355:671–679. https://doi.org/10.1042/bj3550671
Stratilová E, Ait-Mohand F, Řehulka P, Garajová S, Flodrová D, Řehulková H, Farkaš V (2010) Xyloglucan endotransglycosylases (XETs) from germinating nasturtium (Tropaeolum majus) seeds: Isolation and characterization of the major form. Plant Phys Biochem 48:207–215. https://doi.org/10.1016/j.plaphy.2010.01.016
Sulová Z, Baran R, Farkaš V (2003) Divergent modes of action on xyloglucan of two isoenzymes of xyloglucan endo-transglycosylase from Tropaeolum majus. Plant Phys Biochem 41:31–437. https://doi.org/10.1016/S0981-9428(03)00050-0
Thompson JE, Fry SC (2001) Restructuring of wall-bound xyloglucan by transglycosylation in living plant cells. Plant J 26:23–34. https://doi.org/10.1046/j.1365-313x.2001.01005.x
Thompson JE, Smith RC, Fry SC (1997) Xyloglucan undergoes interpolymeric transglycosylation during binding to the plant cell wall in vivo: evidence from 13C/3H dual labelling and isopycnic centrifugation in caesium trifluoroacetate. Biochem J 327:699–708. https://doi.org/10.1042/bj3270699
Vincken JP, York WS, Beldman G, Voragen AGJ (1997) Two general branching patterns of xyloglucan, XXXG and XXGC. Plant Phys 114:9–13. https://doi.org/10.1104/pp.114.1.9
Vissenberg K, Martinez-Vilchez IM, Verbelen JP, Miller JG, Fry SC (2000) In vivo colocalization of xyloglucan endotransglycosylase activity and its donor substrate in the elongation zone of Arabidopsis roots. Plant Cell 12:1229–1237. https://doi.org/10.1105/tpc.12.7.1229
York WS, van Halbeek H, Darvill AG, Albersheim P (1990) Structural analysis of xyloglucan oligosaccharides by 1H-N.M.R. spectroscopy and fast-atom-bombardment mass spectrometry. Carbohydr Res 200:9–31. https://doi.org/10.1016/0008-6215(90)84179-X
York WS, Harvey LK, Guillen R, Albersheim P, Darvill AG (1993) Structural analysis of tamarind seed xyloglucan oligosaccharides using β-galactosidase digestion and spectroscopic methods. Carbohydr Res 248:285–301. https://doi.org/10.1016/0008-6215(93)84135-S
Zemková Z, Garajová S, Flodrová D, Řehulka P, Zelko I, Vadkertiová R, Farkaš V, Stratilová E (2012) Incorporation of β-(1,6)-linked glucooligosaccharides (pustulooligosaccharides) into plant cell wall structures. Chem Pap 66:14–820. https://doi.org/10.2478/s11696-012-0167-x
Acknowledgements
This work was supported by the grant No. 2/0058/16 from VEGA, Slovakia to ES, and by the funding from Huaiyin Normal University and the Australian Research Council Linkage Project (DP120100900) to MH. We thank IBH Wilson from the Universität für Bodenkultur, Vienna, Austria, for providing the pPICZα-His/FLAG plasmid, to I Zelko and R Vadkertiova (Institute of Chemistry) for the assistance with fluorescent microscopy, and H Čigašová (Institute of Chemistry) for technical assistance.
Author information
Authors and Affiliations
Contributions
Conceived, designed experiments and analysed data: B.S., Z.F., J.K., S.Š., E.S. and M.H. Z.F. and J.K. determined the primary structure of TmXET6.3, B.S. and E.S. quantified enzyme activities of wild-type and variants, Á.H., B.S. and F.A-M. run electrophoretic analyses, E.S. conducted microscopy analyses, S.Š. constructed variant plasmids and selected hyper-producing clones, D.S. and S.G. worked out activity assays, S.K. built the 3D homology model, BS conducted large-scale bioinformatics analyses and suggested variant sites, V.F. prepared fluorescent oligosaccharides, M.H. conducted phylogeny reconstruction analyses and generated structural graphics. Discussed the data and contributed to writing: B.S., J.K., S.Š., S.K., V.F., E.S. and M.H. E.S. and M.H. designed research and wrote the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
Authors declare that they have no conflict of interest.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Database accession numbers: The nucleotide sequence of TmXET6.3 is available in GenBank under HF968473, the protein sequence of TmXET6.3 in UniprotKB under V5ZEF7, and the structural data of TmXET6.3 in the Protein Model DataBase under PM0081526.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Stratilová, B., Firáková, Z., Klaudiny, J. et al. Engineering the acceptor substrate specificity in the xyloglucan endotransglycosylase TmXET6.3 from nasturtium seeds (Tropaeolum majus L.). Plant Mol Biol 100, 181–197 (2019). https://doi.org/10.1007/s11103-019-00852-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11103-019-00852-8