Abstract
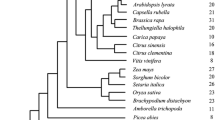
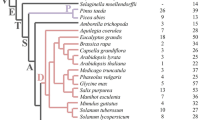
GH3 amino acid conjugases have been identified in many plant and bacterial species. The evolution of GH3 genes in plant species is explored using the sequenced rosids Arabidopsis, papaya, poplar, and grape. Analysis of the sequenced non-rosid eudicots monkey flower and columbine, the monocots maize and rice, as well as spikemoss and moss is included to provide further insight into the origin of GH3 clades. Comparison of co-linear genes in regions surrounding GH3 genes between species helps reconstruct the evolutionary history of the family. Combining analysis of synteny with phylogenetics, gene expression and functional data redefines the Group III GH3 genes, of which AtGH3.12/PBS3, a regulator of stress-induced salicylic acid metabolism and plant defense, is a member. Contrary to previous reports that restrict PBS3 to Arabidopsis and its close relatives, PBS3 syntelogs are identified in poplar, grape, columbine, maize and rice suggesting descent from a common ancestral chromosome dating to before the eudicot/monocot split. In addition, the clade containing PBS3 has undergone a unique expansion in Arabidopsis, with expression patterns for these genes consistent with specialized and evolving stress-responsive functions.






Similar content being viewed by others
Abbreviations
- BTH:
-
1,2,3-benzothiodiazole-7-carbothioic acid S-methyl ester
- IAA:
-
Indole-3-acetic acid
- JA:
-
Jasmonic acid
- SA:
-
Salicylic acid
- bZIP:
-
Basic-domain leucine-zipper
- ERF :
-
Ethylene response factor
- GDG1 :
-
GH3-like defense gene 1
- GH3 :
-
Gretchen Hagen 3
- ICS1 :
-
Isochorismate synthase 1
- JAR1 :
-
Jasmonic acid resistant 1
- PBS3 :
-
avrPphB susceptible 3
- WIN3 :
-
HopW1-1-interacting 3
- Ac:
-
Aquilegia coerulea (columbine)
- At:
-
Arabidopsis thaliana
- Cp:
-
Carica papaya (papaya)
- Mg:
-
Mimulus guttatus (monkey flower)
- Os:
-
Oryza sativa (rice)
- Pp:
-
Physcomitrella patens (moss)
- Pt:
-
Populus trichocarpa (poplar)
- Sm:
-
Selaginella moellendorffii (spikemoss)
- Vv:
-
Vitis vinifera (grape)
- Zm:
-
Zea mays (maize)
- ML:
-
Maximum likelihood
- MP:
-
Maximum parsimony
- NJ:
-
Neighbor joining
- PID:
-
Percent identity
- WGD:
-
Whole genome duplication
References
Albertazzi G, Milc J, Caffagni A, Francia E, Roncaglia E et al (2009) Gene expression in grapevine cultivars in response to Bois noir phytoplasma infection. Plant Sci 176:792–804
Ariizumi T, Hatakeyama K, Hinata K, Inatsugi R, Nishida I, Sata S, Kato T, Tabata S, Toriyama K (2004) Disruption of the novel plant protein NEF1 affects lipid accumulation in the plastids of the tapetum and exine formation of pollen, resulting in male sterility in Arabidopsis thaliana. Plant J 39:170–181
Bierfreund NM, Tintelnot S, Reski R, Decker EL (2004) Loss of GH3 function does not affect phytochrome-mediated development in a moss, Physcomitrella patens. J Plant Physiol 161:823–835
Blanc G, Hokamp K, Wolfe KH (2003) A recent polyploidy superimposed on older large-scale duplications in the Arabidopsis genome. Genome Res 13:137–144
Bourne DJ, Barrow KD, Milborrow BV (1991) Salicyloylaspartate as an endogenous component of the leaves of Phaseolus vulgaris. Phytochemistry 30:4041–4044
Castresana J (2000) Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol 17:540–552
Chang K-H, Xiang H, Dunaway-Mariano D (1997) Acyl-adenylate motif of the acyl-adenylate/thioester-forming enzyme superfamily: a site directed mutagenesis study with the Pseudomonas sp. Strain CBS3 4-chlorobenzoate:Coenzyme A ligase. Biochemistry 36:15650–15659
Chini A, Fonseca S, Fernandez G, Adie B, Chico JM et al (2007) The JAZ family of repressors is the missing link in jasmonate signalling. Nature 448:666
Conti E, Franks NP, Brick P (1996) Crystal structure of firefly luciferase throws light on a superfamily of adenylate-forming enzymes. Structure 4:287–298
Craigon DJ, James N, Okyere J, Higgins J, Jotham J et al (2004) Nascarrays: a repository for microarray data generated by NASC’s transcriptomics service. Nucleic Acids Res 32:D575–D577
Dereeper A, Guignon V, Blanc G, Audic S, Buffet S et al (2008) Phylogeny.Fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res 36:W465–W469
Ding X, Cao Y, Huang L, Zhao J, Xu C et al (2008) Activation of the indole-3-acetic acid-amido synthetase GH3–8 suppresses expansin expression and promotes salicylate- and jasmonate-independent basal immunity in rice. Plant Cell 20:228–240
Dombrecht B, Xue GP, Sprague SJ, Kirkegaard JA, Ross JJ et al (2007) MYC2 differentially modulates diverse jasmonate-dependent functions in Arabidopsis. Plant Cell 19:2225–2245
Domingo C, Andres F, Tharreau D, Iglesias DJ, Talon M (2009) Constitutive expression of OsGH3.1 reduces auxin content and enhances defense response and resistance to a fungal pathogen in rice. Mol Plant-Micro Interact 22:201–210
Dudareva N, Pichersky E (2000) Biochemical and molecular genetic aspects of floral scents. Plant Physiol 122:627–634
Edgar RC (2004) Muscle: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32:1792–1797
Felten J, Kohler A, Morin E, Bhalerao RP, Palme K et al (2009) The ectomycorrhizal fungus Laccaria bicolor stimulates lateral root formation in poplar and Arabidopsis through auxin transport and signaling. Plant Physiol 151:1991–2005
Ferrari S, Plotnikova JM, De Lorenzo G, Ausubel FM (2003) Arabidopsis local resistance to Botrytis cinerea involves salicylic acid and camalexin and requires EDS4 and PAD2, but not SID2, EDS5, or PAD4. Plant J 35:193–205
Freeling M (2009) Bias in plant gene content following different sorts of duplication: Tandem, whole-genome, segmental, or by transposition. Annu Rev Plant Biol 60:433–453
Goda H, Sasaki E, Akiyama K, Maruyama-Nakashita A, Nakabayashi K et al (2008) The AtGenExpress hormone and chemical treatment data set: experimental design, data evaluation, model data analysis and data access. Plant J 55:526–542
Guindon S, Gascuel O (2003) A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol 52:696–704
Gutterson N, Reuber TL (2004) Regulation of disease resistance pathways by AP2/ERF transcription factors. Curr Opin Plant Biol 7:465–471
Haberer G, Hindemitt T, Meyers BC, Mayer KFX (2004) Transcriptional similarities, dissimilarities, and conservation of cis-elements in duplicated genes of Arabidopsis. Plant Physiol 136:3009–3022
Hagen G, Kleinschmidt A, Guilfoyle T (1984) Auxin-regulated gene expression in intact soybean Glycine max cultivar Wayne hypocotyl and excised hypocotyl sections. Planta 162:147–153
Hall BG (2005) Comparison of the accuracies of several phylogenetic methods using protein and DNA sequences. Mol Biol Evol 22:792–802
Hanada K, Zou C, Lehti-Shiu MD, Shinozaki K, Shiu SH (2008) Importance of lineage-specific expansion of plant tandem duplicates in the adaptive response to environmental stimuli. Plant Physiol 148:993–1003
Hsieh H-L, Okamoto H, Wang M, Ang L-H, Matsui M et al (2000) FIN219, an auxin-regulated gene, defines a link between phytochrome A and the downstream regulator COP1 in light control of Arabidopsis development. Genes Dev 14:1958–1970
Huelsenbeck JP, Hillis DM (1993) Success of phylogenetic methods in the four-taxon case. Syst Biol 42:247–264
Jagadeeswaran G, Raina S, Acharya BR, Maqbool SB, Mosher SL et al (2007) Arabidopsis GH3-like Defense Gene 1 is required for accumulation of salicylic acid, activation of defense responses and resistance to Pseudomonas syringae. Plant J 51:234–246
Jaillon O, Aury JM, Noel B, Policriti A, Clepet C et al (2007) The grapevine genome sequence suggests ancestral hexaploidization in major angiosperm phyla. Nature 449:463–467
Jain M, Kaur N, Tyagi AK, Khurana JP (2006) The auxin-responsive GH3 gene family in rice (oryza sativa). Funct Integr Genomics 6:36–46
Jansen R, Kaittanis C, Saski C, Lee S-B, Tomkins J et al (2006) Phylogenetic analyses of Vitis (Vitaceae) based on complete chloroplast genome sequences: effects of taxon sampling and phylogenetic methods on resolving relationships among rosids. BMC Evol Biol 6:32
Jun J, Mandoiu I, Nelson C (2009) Identification of mammalian orthologs using local synteny. BMC Genomics 10:630
Kessler D, Diezel C, Baldwin IT (2010) Changing pollinators as a means of escaping herbivores. Curr Biol 20:237–242
Khan S, Stone JM (2007) Arabidopsis thaliana GH3.9 influences primary root growth. Planta 226:21–34
Krishnaswamy S, Verma S, Rahman MH, Kav NNV (2011) Functional characterization of four APETALA2-family genes (RAP2.6, RAP2.6L, DREB19 and DREB26) in Arabidopsis. Plant Mol Biol 75:107–127
Lee MW, Lu H, Jung HW, Greenberg JT (2007) A key role for the Arabidopsis WIN3 protein in disease resistance triggered by Pseudomonas syringae that secrete AvrRpt2. MPMI 20:1192–1200
Lee MW, Jelenska J, Greenberg JT (2008) Arabidopsis proteins important for modulating defense responses to Pseudomonas syringae that secrete HopW1–1. Plant J 54:452–465
Liu Kl, Kang B-C, Jiang H, Moore SL, Li H et al (2005) A GH3-like gene, CcGH3, isolated from Capsicum chinense l. Fruit is regulated by auxin and ethylene. Plant Mol Biol 58:447–464
Ludwig-Muller J, Julke S, Bierfreund NM, Decker EL, Reski R (2009) Moss (Physcomitrella patens) GH3 proteins act in auxin homeostasis. New Phytol 181:323–338
Lyons E, Freeling M (2008) How to usefully compare homologous plant genes and chromosomes as DNA sequences. Plant J 53:661–673
Lyons E, Pedersen B, Kane J, Alam M, Ming R et al (2008) Finding and comparing syntenic regions among Arabidopsis and the outgroups papaya, poplar, and grape: CoGe with rosids. Plant Physiol 148:1772–1781
Martinez C, Pons E, Prats G, Leon J (2004) Salicylic acid regulates flowering time and links defence responses and reproductive development. Plant J 37:209–217
Ming R, Hou S, Feng Y, Yu Q, Dionne-Laporte A et al (2008) The draft genome of the transgenic tropical fruit tree papaya (Carica papaya linnaeus). Nature 452:991–996
Molina C, Grotewold E (2005) Genome wide analysis of Arabidopsis core promoters. BMC Genomics 6:25
Mount DW (2004) Bioinformatics: sequence and genome analysis. Cold Spring Harbor Laboratory Press, Cold Spring Harbor
Nakazawa M, Yabe N, Ichikawa T, Yamamoto YY, Yoshizumi T et al (2001) DFL1, an auxin-responsive GH3 gene homologue, negatively regulates shoot cell elongation and lateral root formation, and positively regulates the light response of hypocotyl length. Plant J 25:213–221
Nobuta K, Okrent RA, Stoutemyer M, Rodibaugh N, Kempema L et al (2007) The GH3 acyl adenylase family member PBS3 regulates salicylic acid-dependent defense responses in Arabidopsis. Plant Physiol 144:1144–1156
Okrent RA, Brooks MD, Wildermuth MC (2009) Arabidopsis GH3.12 (PBS3) conjugates amino acids to 4-substituted benzoates and is inhibited by salicylate. J Biol Chem 284:9742–9754
Ouyang S, Zhu W, Hamilton J, Lin H, Campbell M et al (2006) The TIGR rice genome annotation resource: improvements and new features. Nucleic Acids Res 35:D883–D887
Park J-E, Seo PJ, Lee A-K, Jung J-H, Kim Y-S et al (2007a) An Arabidopsis GH3 gene, encoding an auxin-conjugating enzyme, mediates phytochrome B-regulated light signals in hypocotyl growth. Plant Cell Physiol 48:1236–1241
Park J-E, Park J-Y, Kim Y-S, Staswick PE, Jeon J, Yun J, Kim S-Y, Kim J, Lee Y-H, Park C-M (2007b) GH3-mediated auxin homeostasis links growth regulation with stress adaptation response in Arabidopsis. J Biol Chem 282:10036–10046
Rensing SA, Lang D, Zimmer AD, Terry A, Salamov A et al (2008) The Physcomitrella genome reveals evolutionary insights into the conquest of land by plants. Science 319:64–69
Rizzon C, Ponger L, Gaut BS (2006) Striking similarities in the genomic distribution of tandemly arrayed genes in Arabidopsis and rice. PLoS Comput Biol 2:e115
Schnable PS, Ware D, Fulton RS, Stein JC, Wei F et al (2009) The B73 maize genome: complexity, diversity, and dynamics. Science 326:1112–1115
Semon M, Wolfe KH (2007) Consequences of genome duplication. Curr Opin Genet Dev 17:505–512
Singh KB, Foley RC, Onate-Sanchez L (2002) Transcription factors in plant defense and stress responses. Curr Opin Plant Biol 5:430–436
Staswick PE, Tiryaki I (2004) The oxylipin signal jasmonic acid is activated by an enzyme that conjugates it to isoleucine in Arabidopsis. Plant Cell 16:2117–2127
Staswick PE, Su W, Howell SH (1992) Methyl jasmonate inhibition of root growth and induction of a leaf protein are decreased in an Arabidopsis thaliana mutant. Proc Natl Acad Sci USA 89:6837–6840
Staswick PE, Tiryaki I, Rowe ML (2002) Jasmonate response locus JAR1 and several related Arabidopsis genes encode enzymes of the firefly luciferase superfamily that show activity on jasmonic, salicylic, and indole-3-acetic acids in an assay for adenylation. Plant Cell 14:1405–1415
Staswick PE, Serban B, Rowe M, Tiryaki I, Maldonado MT et al (2005) Characterization of an Arabidopsis enzyme family that conjugates amino acids to indole-3-acetic acid. Plant Cell 17:616–627
Steel M (2005) Should phylogenetic models be trying to “Fit an elephant”? Trends Genet 21:307–309
Sun F, Liu P, Xu J, Dong H (2010) Mutation in RAP2.6L, a transactivator of the ERF transcription factor family enhances resistance to Pseudomonas syringae. Physiol Mol Plant Pathol 74:295–301
Suzuki Y, Yanaguchi I, Murofushi N, Takahasi N (1988) Biological conversion of benzoic acid in Lemna paucicostata 151 and its relation to flower induction. Plant Cell Physiol 29:439–444
Takase T, Nakazawa M, Ishikawa A, Manabe K, Matsui M (2003) DFL2, a new member of the Arabidopsis GH3 family is involved in red light-specific hypocotyl elongation. Plant Cell Physiol 44:1071–1080
Takase T, Nakazawa M, Ishikawa A, Kawashima M, Ichikawa T, Takahashi N, Shimada H, Manabe K, Matsui M (2004) ydk1-D, an auxin-responsive GH3 mutant that is involved in hypocotyl and root elongation. Plant J 37:471–483
Tamura K, Dudley J, Nei M, Kumar S (2007) MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol 24:1596–1599
Terol J, Domingo C, Talon M (2006) The GH3 family in plants: genome wide analysis in rice and evolutionary history based on EST analysis. Gene 371:279–290
Thornton JW, Kolaczkowski B (2005) No magic pill for phylogenetic error. Trends Genet 21:310–311
Toufighi K, Brady SM, Austin R, Ly E, Provart NJ (2005) The botany array resource: E-northerns, expression angling, and promoter analyses. Plant J 43:153–163
Trennheuser F, Burkhard G, Becker H (1994) Anthocerodiazonin, an alkaloid from Antheroceros agretis. Phytochemistry 37:899–903
Tuskan GA, DiFazio S, Jansson S, Bohlmann J, Grigoriev I et al (2006) The genome of black cottonwood, populus trichocarpa (torr. & gray). Science 313:1596–1604
Warren RF, Merritt PM, Holub E, Innes RW (1999) Identification of three putative signal transduction genes involved in R gene-specified disease resistance in Arabidopsis. Genetics 152:401–412
Winter D, Vinegar B, Nahal H, Ammar R, Wilson GV et al (2007) An “electronic fluorescent pictograph” browser for exploring and analyzing large-scale biological data sets. PLoS One 2:e718
Wise RP, Caldo RA, Hong L, Shen L, Cannon E et al (2007) Barleybase/plexdb: a unified expression profiling database for plants and plant pathogens. In: Edwards D (ed) Plant bioinformatics—methods and protocols. Humana Press, Totowa, pp 347–363
Woodward AW, Bartel B (2005) Auxin: regulation, action, and interaction. Ann Bot 95:707–735
Wu CA, Lowry DB, Cooley AM, Wright KM, Lee YW et al (2007) Mimulus is an emerging model system for the integration of ecological and genomic studies. Heredity 100:220–230
Zhang Z, Li Q, Li Z, Staswick PE, Wang M et al (2007) Dual regulation role of GH3.5 in salicylic acid and auxin signaling during Arabidopsis-Pseudomonas syringae interaction. Plant Physiol 145:450–464
Zhang S-W, Li C-H, Cao J, Zhang Y-C, Zhang S-Q et al (2009) Altered architecture and enhanced drought tolerance in rice via the down-regulation of indole-3-acetic acid by tld1/osgh3.13 activation. Plant Physiol 151:1889–1901
Zimmermann P, Hirsch-Hoffmann M, Hennig L, Gruissem W (2004) GENEVESTIGATOR. Arabidopsis microarray database and analysis toolbox. Plant Physiol 136:2621–2632
Acknowledgments
We would like to thank Dr. Eric Lyons for his assistance with the CoGe browser, Dr. Divya Chandran for careful reading of the manuscript, and the William Carroll Smith Graduate Research Fellowship in Plant Pathology (to R.A.O) and UC Berkeley awards (to M.C.W.) for financial support. Some of the genome sequence data described here was analyzed prior to publication by the sequencing projects. Of these, the Aquilegia coerulea, Mimulus guttatus, and Selaginella moellendorffii data were produced by the US Department of Energy Joint Genome Institute. Carica papaya data were produced by the ASGPB Hawaii Papaya Genome Project (http://www.asgpb.mhpcc.hawaii.edu/papaya/). Zea mays data were produced by the Genome Sequencing Center at Washington University School of Medicine in St. Louis and can be obtained from http://www.maizesequence.org/.
Author information
Authors and Affiliations
Corresponding author
Additional information
Accession numbers: AtGH3.1, At2g14960; AtGH3.2, At4g37390; AtGH3.3, At2g23170; AtGH3.4, At1g59500; AtGH3.5, At4g27260; AtGH3.6, At5g54510; AtGH3.7, At1g23160; AtGH3.8, At5g51470; AtGH3.9, At2g47750; AtGH3.10, At4g03400; AtGH3.11, At2g46370; AtGH3.12, At5g13320; AtGH3.13, At5g13350; AtGH3.14, At5g13360; AtGH3.15, At5g13370; AtGH3.16, At5g13380; AtGH3.17, At1g28130; AtGH3.18, At1g48670; AtGH3.19, At1g48660.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Okrent, R.A., Wildermuth, M.C. Evolutionary history of the GH3 family of acyl adenylases in rosids. Plant Mol Biol 76, 489–505 (2011). https://doi.org/10.1007/s11103-011-9776-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11103-011-9776-y