Abstract
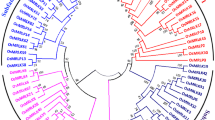
X1-homologous genes (XHS) encode plant-specific proteins containing three major domains (XH, XS, zf-XS), but their functions are largely unknown. We report the systematic identification and characterization of XHS genes in the rice genome. Eleven putative XHS protein sequences (OXHS1–11) were identified in the sequenced genome of Oryza sativa japonica cv. Nipponbare, and these sequences, along with other plant XHS homologues, were classified into five subgroups based on phylogenetic analysis. Distinct diversification of the XHS proteins occurred between monocotyledon and dicotyledon plants. The OXHS family has diverse exon–intron structures and organizations of putative domains and motifs. The OXHS proteins showed no transactivation activity, and no interaction between the XH domain and the XS domain in yeast. Four representative OXHS proteins were targeted to cytoplasm, which contradicts the previous speculation that XHS proteins are putative transcription factors. All the OXHS genes are predominantly expressed in floral organs, and some are expressed in a wide range of tissues or organs in indica rice Minghui 63. Nine OXHS genes are responsive to at least one of the abiotic stresses including drought, salt, cold, and abscisic acid treatment. Over-expression of one stress-responsive gene OXHS2 in rice resulted in reduced tolerance to salt and drought stresses. These results suggest that the OXHS family may be functionally diversified and some members of this family may play important roles in regulating stress tolerance in rice.








Similar content being viewed by others
Abbreviations
- ABA:
-
Abscisic acid
- XH:
-
X1 homologue
- XHS:
-
XH and XS domain
- PCR:
-
Polymerase chain reaction
- RT:
-
Reverse transcription
References
Bateman A (2002) The SGS3 protein involved in PTGS finds a family. BMC Bioinformatics 3:21
Bateman A, Coin L, Durbin R, Finn RD, Hollich V, Griffiths-Jones S, Khanna A, Marshall M, Moxon S, Sonnhammer EL, Studholme DJ, Yeats C, Eddy SR (2004) The Pfam protein families database. Nucleic Acids Res 32:D138–D141
Boulikas T (1994) Putative nuclear localization signals (NLS) in protein transcription factors. J Cell Biochem 55:32–58
Calderon-Villalobos LI, Nill C, Marrocco K, Kretsch T, Schwechheimer C (2007) The evolutionarily conserved Arabidopsis thaliana F-box protein AtFBP7 is required for efficient translation during temperature stress. Gene 392:106–116
Chae HJ, Park JM, Lee GY, Park HR, Chae SW, Jeong GS, Kim HM, Kim SB, Yoo SK, Kim HR (2004) Yuk-Hap-Tang induces apoptosis by intervening mn-SOD in human cervical carcinoma HeLa cells. Am J Chin Med 32:883–895
Chen M, Bennetzen JL (1996) Sequence composition and organization in the Sh2/A1-homologous region of rice. Plant Mol Biol 32:999–1001
Dai X, Xu Y, Ma Q, Xu W, Wang T, Xue Y, Chong K (2007) Overexpression of an R1R2R3 MYB gene, OsMYB3R–2, increases tolerance to freezing, drought, and salt stress in transgenic Arabidopsis. Plant Physiol 143:1739–1751
Durfee T, Roe JL, Sessions RA, Inouye C, Serikawa K, Feldmann KA, Weigel D, Zambryski PC (2003) The F-box-containing protein UFO and AGAMOUS participate in antagonistic pathways governing early petal development in Arabidopsis. Proc Natl Acad Sci U S A 100:8571–8576
Fraser CM, Rider LW, Chapple C (2005) An expression and bioinformatics analysis of the Arabidopsis serine carboxypeptidase-like gene family. Plant Physiol 138:1136–1148
Gasteiger E, Gattiker A, Hoogland C, Ivanyi I, Appel RD, Bairoch A (2003) ExPASy: the proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res 31:3784–3788
Glick E, Zrachya A, Levy Y, Mett A, Gidoni D, Belausov E, Citovsky V, Gafni Y (2008) Interaction with host SGS3 is required for suppression of RNA silencing by tomato yellow leaf curl virus V2 protein. Proc Natl Acad Sci U S A 105:157–161
Goldfarb AN, Lewandowska K (1994) Nuclear redirection of a cytoplasmic helix-loop-helix protein via heterodimerization with a nuclear localizing partner. Exp Cell Res 214:481–485
Guiltinan MJ, Miller L (1994) Molecular characterization of the DNA-binding and dimerization domains of the bZIP transcription factor, EmBP-1. Plant Mol Biol 26:1041–1053
He XJ, Mu RL, Cao WH, Zhang ZG, Zhang JS, Chen SY (2005) AtNAC2, a transcription factor downstream of ethylene and auxin signaling pathways, is involved in salt stress response and lateral root development. Plant J 44:903–916
Hegedus D, Yu M, Baldwin D, Gruber M, Sharpe A, Parkin I, Whitwill S, Lydiate D (2003) Molecular characterization of Brassica napus NAC domain transcriptional activators induced in response to biotic and abiotic stress. Plant Mol Biol 53:383–397
Hiei Y, Ohta S, Komari T, Kumashiro T (1994) Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T-DNA. Plant J 6:271–282
Hu H, Dai M, Yao J, Xiao B, Li X, Zhang Q, Xiong L (2006) Overexpressing a NAM, ATAF, and CUC (NAC) transcription factor enhances drought resistance and salt tolerance in rice. Proc Natl Acad Sci U S A 103:12987–12992
Huala E, Dickerman AW, Garcia-Hernandez M, Weems D, Reiser L, LaFond F, Hanley D, Kiphart D, Zhuang M, Huang W, Mueller LA, Bhattacharyya D, Bhaya D, Sobral BW, Beavis W, Meinke DW, Town CD, Somerville C, Rhee SY (2001) The Arabidopsis Information Resource (TAIR): a comprehensive database and web-based information retrieval, analysis, and visualization system for a model plant. Nucleic Acids Res 29:102–105
Huang H, Tudor M, Su T, Zhang Y, Hu Y, Ma H (1996) DNA binding properties of two Arabidopsis MADS domain proteins: binding consensus and dimer formation. Plant Cell 8:81–94
Imai N, Matsuda N, Tanaka K, Nakano A, Matsumoto S, Kang W (2003) Ubiquitin ligase activities of Bombyx mori nucleopolyhedrovirus RING finger proteins. J Virol 77:923–930
Itoh J, Nonomura K, Ikeda K, Yamaki S, Inukai Y, Yamagishi H, Kitano H, Nagato Y (2005) Rice plant development: from zygote to spikelet. Plant Cell Physiol 46:23–47
Jain M, Nijhawan A, Arora R, Agarwal P, Ray S, Sharma P, Kapoor S, Tyagi AK, Khurana JP (2007) F-box proteins in rice. Genome-wide analysis, classification, temporal and spatial gene expression during panicle and seed development, and regulation by light and abiotic stress. Plant Physiol 143:1467–1483
Joazeiro CA, Weissman AM (2000) RING finger proteins: mediators of ubiquitin ligase activity. Cell 102:549–552
Kikuchi S, Satoh K, Nagata T, Kawagashira N, Doi K, Kishimoto N, Yazaki J, Ishikawa M, Yamada H, Ooka H, Hotta I, Kojima K, Namiki T, Ohneda E, Yahagi W, Suzuki K, Li CJ, Ohtsuki K, Shishiki T, Otomo Y, Murakami K, Iida Y, Sugano S, Fujimura T, Suzuki Y, Tsunoda Y, Kurosaki T, Kodama T, Masuda H, Kobayashi M, Xie Q, Lu M, Narikawa R, Sugiyama A, Mizuno K, Yokomizo S, Niikura J, Ikeda R, Ishibiki J, Kawamata M, Yoshimura A, Miura J, Kusumegi T, Oka M, Ryu R, Ueda M, Matsubara K, Kawai J, Carninci P, Adachi J, Aizawa K, Arakawa T, Fukuda S, Hara A, Hashizume W, Hayatsu N, Imotani K, Ishii Y, Itoh M, Kagawa I, Kondo S, Konno H, Miyazaki A, Osato N, Ota Y, Saito R, Sasaki D, Sato K, Shibata K, Shinagawa A, Shiraki T, Yoshino M, Hayashizaki Y, Yasunishi A (2003) Collection, mapping, and annotation of over 28, 000 cDNA clones from japonica rice. Science 301:376–379
Koiwa H, Barb AW, Xiong L, Li F, McCully MG, Lee BH, Sokolchik I, Zhu J, Gong Z, Reddy M, Sharkhuu A, Manabe Y, Yokoi S, Zhu JK, Bressan RA, Hasegawa PM (2002) C-terminal domain phosphatase-like family members (AtCPLs) differentially regulate Arabidopsis thaliana abiotic stress signaling, growth, and development. Proc Natl Acad Sci U S A 99:10893–10898
Liu L, White MJ, MacRae TH (1999) Transcription factors and their genes in higher plants functional domains, evolution and regulation. Eur J Biochem 262:247–257
Lupas AN, Gruber M (2005) The structure of alpha-helical coiled coils. Adv Protein Chem 70:37–78
Lupas A, Van Dyke M, Stock J (1991) Predicting coiled coils from protein sequences. Science 252:1162–1164
Marmorstein R, Fitzgerald MX (2003) Modulation of DNA-binding domains for sequence-specific DNA recognition. Gene 304:1–12
Mitsuda N, Iwase A, Yamamoto H, Yoshida M, Seki M, Shinozaki K, Ohme-Takagi M (2007) NAC transcription factors, NST1 and NST3, are key regulators of the formation of secondary walls in woody tissues of Arabidopsis. Plant Cell 19:270–280
Mittler R (2006) Abiotic stress, the field environment and stress combination. Trends Plant Sci 11:15–19
Mourrain P, Beclin C, Elmayan T, Feuerbach F, Godon C, Morel JB, Jouette D, Lacombe AM, Nikic S, Picault N, Remoue K, Sanial M, Vo TA, Vaucheret H (2000) Arabidopsis SGS2 and SGS3 genes are required for posttranscriptional gene silencing and natural virus resistance. Cell 101:533–542
Muangsan N, Beclin C, Vaucheret H, Robertson D (2004) Geminivirus VIGS of endogenous genes requires SGS2/SDE1 and SGS3 and defines a new branch in the genetic pathway for silencing in plants. Plant J 38:1004–1014
Ohnishi T, Sugahara S, Yamada T, Kikuchi K, Yoshiba Y, Hirano HY, Tsutsumi N (2005) OsNAC6, a member of the NAC gene family, is induced by various stresses in rice. Genes Genet Syst 80:135–139
Page RD (1996) TreeView: an application to display phylogenetic trees on personal computers. Comput Appl Biosci 12:357–358
Peragine A, Yoshikawa M, Wu G, Albrecht HL, Poethig RS (2004) SGS3 and SGS2/SDE1/RDR6 are required for juvenile development and the production of trans-acting siRNAs in Arabidopsis. Genes Dev 18:2368–2379
Ronquist F, Huelsenbeck JP (2003) MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19:1572–1574
Rose A, Schraegle SJ, Stahlberg EA, Meier I (2005) Coiled-coil protein composition of 22 proteomes–differences and common themes in subcellular infrastructure and traffic control. BMC Evol Biol 5:66
Rotman N, Durbarry A, Wardle A, Yang WC, Chaboud A, Faure JE, Berger F, Twell D (2005) A novel class of MYB factors controls sperm-cell formation in plants. Curr Biol 15:244–248
Saijo Y (2000) Over-expression of a single Ca2+-dependent protein kinase confers both cold and salt/drought tolerance on rice plants. Plant J 23:319–327
Sainz MB, Goff SA, Chandler VL (1997a) Extensive mutagenesis of a transcriptional activation domain identifies single hydrophobic and acidic amino acids important for activation in vivo. Mol Cell Biol 17:115–122
Sainz MB, Grotewold E, Chandler VL (1997b) Evidence for direct activation of an anthocyanin promoter by the maize C1 protein and comparison of DNA binding by related Myb domain proteins. Plant Cell 9:611–625
Schwechheimer C, Smith C, Bevan MW (1998) The activities of acidic and glutamine-rich transcriptional activation domains in plant cells: design of modular transcription factors for high-level expression. Plant Mol Biol 36:195–204
Spittau B, Wang Z, Boinska D, Krieglstein K (2007) Functional domains of the TGF-beta-inducible transcription factor Tieg3 and detection of two putative nuclear localization signals within the zinc finger DNA-binding domain. J Cell Biochem 101:712–722
Tominaga R, Iwata M, Sano R, Inoue K, Okada K, Wada T (2008) Arabidopsis CAPRICE-LIKE MYB 3 (CPL3) controls endoreduplication and flowering development in addition to trichome and root hair formation. Development 135:1335–1345
Urao T, Yamaguchi-Shinozaki K, Urao S, Shinozaki K (1993) An Arabidopsis myb homolog is induced by dehydration stress and its gene product binds to the conserved MYB recognition sequence. Plant Cell 5:1529–1539
Van de Graaff WB, Thompson WL, Sunshine I, Fretthold D, Leickly F, Dayton H (1982) Adsorbent and cathartic inhibition of enteral drug absorption. J Pharmacol Exp Ther 221:656–663
Washburn KB, Davis EA, Ackerman S (1997) Coactivators and TAFs of transcription activation in wheat. Plant Mol Biol 35:1037–1043
Wolf E, Kim PS, Berger B (1997) MultiCoil: a program for predicting two- and three-stranded coiled coils. Protein Sci 6:1179–1189
Woo HR, Chung KM, Park JH, Oh SA, Ahn T, Hong SH, Jang SK, Nam HG (2001) ORE9, an F-box protein that regulates leaf senescence in Arabidopsis. Plant Cell 13:1779–1790
Xiao B, Chen X, Xiang C, Tang N, Zhang Q, Xiong L (2009) Evaluation of seven function-known candidate genes for their effects on improving drought resistance of transgenic rice under field conditions. Mol Plant 2:73–83
Xie K, Wu C, Xiong L (2006) Genomic organization, differential expression, and interaction of SQUAMOSA promoter-binding-like transcription factors and microRNA156 in rice. Plant Physiol 142:280–293
Yamaguchi M, Kubo M, Fukuda H, Demura T (2008) Vascular-related NAC-DOMAIN7 is involved in the differentiation of all types of xylem vessels in Arabidopsis roots and shoots. Plant J 55:652–664
Yamasaki K, Kigawa T, Inoue M, Watanabe S, Tateno M, Seki M, Shinozaki K, Yokoyama S (2008) Structures and evolutionary origins of plant-specific transcription factor DNA-binding domains. Plant Physiol Biochem 46:394–401
Yanagisawa S, Sheen J (1998) Involvement of maize Dof zinc finger proteins in tissue-specific and light-regulated gene expression. Plant Cell 10:75–89
Yu YB (2002) Coiled-coils: stability, specificity, and drug delivery potential. Adv Drug Deliv Rev 54:1113–1129
Yuan Q, Ouyang S, Wang A, Zhu W, Maiti R, Lin H, Hamilton J, Haas B, Sultana R, Cheung F, Wortman J, Buell CR (2005) The institute for genomic research Osa1 rice genome annotation database. Plant Physiol 138:18–26
Yue B, Xue W, Xiong L, Yu X, Luo L, Cui K, Jin D, Xing Y, Zhang Q (2006) Genetic basis of drought resistance at reproductive stage in rice: separation of drought tolerance from drought avoidance. Genetics 172:1213–1228
Zhu QH, Ramm K, Shivakkumar R, Dennis ES, Upadhyaya NM (2004) The ANTHER INDEHISCENCE1 gene encoding a single MYB domain protein is involved in anther development in rice. Plant Physiol 135:1514–1525
Acknowledgments
This work was supported by grants from the National Special Key Project of China on Functional Genomics of Major Plants and Animals, the National Natural Science Foundation of China, and the Ministry of Education of China.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Qin, Y., Ye, H., Tang, N. et al. Systematic identification of X1-homologous genes reveals a family involved in stress responses in rice. Plant Mol Biol 71, 483 (2009). https://doi.org/10.1007/s11103-009-9535-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11103-009-9535-5