Abstract
In intact plants, cells in axillary buds are arrested at the G1 phase of the cell cycle during dormancy. In mammalian cells, the cell cycle is suppressed at the G1 phase by the activities of retinoblastoma tumor suppressor gene (RB) family proteins, depending on their phosphorylation state. Here, we report the isolation of a pea cDNA clone encoding an RB-related protein (PsRBR1, Accession No. AB012024) with a high degree of amino acid conservation in comparison with RB family proteins. PsRBR1 protein was detected as two polypeptides using an anti-PsRBR1 antibody in dormant axillary buds, whereas it was detected as three polypeptides, which were the same two polypeptides and another larger polypeptide 2 h after terminal decapitation. Both in vitro-synthesized PsPRB1 protein and lambda protein phosphatase-treated PsRBR1 protein corresponded to the smallest polypeptide detected by anti-PsRBR1 antibody, suggesting that the three polypeptides correspond to non-phosphorylated form of PsRBR1 protein, and lower- and higher-molecular mass forms of phosphorylated PsRBR1 protein. Furthermore, in vivo labeling with [32P]-inorganic phosphate indicated that PsRBR1 protein was more phosphorylated before mRNA accumulation of cell cycle regulatory genes such as PCNA. Together these findings suggest that dormancy-to-growth transition in pea axillary buds is regulated by molecular mechanisms of cell cycle control similar to those in mammals, and that the PsRBR1 protein has an important role in suppressing the cell cycle during dormancy in axillary buds.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
In intact plants, the terminal bud predominantly grows, while axillary bud growth is suppressed. This phenomenon is called apical dominance. After terminal bud decapitation, however, the axillary buds grow rapidly and develop in the same manner as the terminal bud. Apical dominance was one of the first developmental phenomena shown to be regulated by plant hormones (Thimann and Skoog 1934). The outgrowth of axillary buds is inhibited by auxin flow from the terminal bud (Booker et al. 2003), and is promoted by cytokinin biosynthesized in the stem after decapitation (Tanaka et al. 2006). Recently, a variety of experimental approaches were used to examine the molecular mechanisms that control shoot branching (Bennett and Leyser 2006; Dun et al. 2006; Shimizu-Sato and Mori 2001). Some mutants were isolated and characterized in Arabidopsis (max1, 2, 3, and 4), pea (rms1, 2, 3, 4, and 5), and petunia (dad1 and 2). Findings from these molecular genetic approaches combined with conventional physiologic studies, such as grafting experiments, suggested that novel signals regulate axillary bud dormancy. The molecular mechanisms of cell cycle control in axillary buds, however, are still not well understood.
Most cells in pea axillary buds are arrested at the G1 phase in the cell cycle, and axillary buds have the capacity to undergo multiple cycles of dormancy and growth (Devitt and Stafstrom 1995; Shimizu and Mori 1998). Arrest and progression of the cell cycle in axillary buds are controlled in response to environmental signals and by developmental programs. Thus, we hypothesized that cells in dormant axillary buds are regulated through suppression mechanisms of cell cycle control suppression mechanisms similar to those in mammals.
In mammals, arrest and progression of the cell cycle are also controlled during the G1 phase in response to environmental signals and by developmental programs (Sherr 2004). This process is governed by both positive- and negative-regulatory factors. One of the most important key regulators is the product of the retinoblastoma tumor suppressor gene (RB). The human RB gene encodes a 110-kDa nuclear phosphoprotein (pRB). pRB activities are regulated by phosphorylation in a cell cycle-dependent manner. Phosphorylation states of pRB regulate the interactions of multiple cellular proteins. In the G1 phase, hypo-phosphorylated pRB (hypo-ppRB) binds to a transcriptional factor, E2F family protein, and suppresses its transcriptional activity. E2F family proteins regulate the transcription of several genes (e.g., dihydrofolate reductase, thymidine kinase, DNA polymerase α, and proliferating cell nuclear antigen [PCNA]) whose products are required for either G1/S transition or DNA replication. Thus, by negatively regulating E2F family proteins, pRB negatively controls cell cycle progression. pRB is highly phosphorylated by several types of cyclin and cyclin-dependent kinase (CDK) complexes in response to environmental signals or developmental programs to progress the cell cycle. E2F family proteins are released from hyper-ppRB and promote the transcription of genes for G1/S transition or DNA replication. Thus, pRB regulates the arrest and progression of the cell cycle based on its phosphorylation state.
In mammals, there are pRB-related proteins, p130 and p107 (Graña et al. 1998). RB family proteins regulate the progression of the cell cycle as well as the entry/exit transition of the cell cycle. RB family proteins have the same pocket protein feature; that is, they interact with a multitude of cellular proteins in a phosphorylation-dependent manner. RB family proteins are, however, modulated by differential regulation of phosphorylation states and protein levels in quiescent cells re-entering the cell cycle. In quiescent cells and in cells at the G0/G1 transition, pRB is in a hypo-phosphorylated form, and p130 is detected in non- and low-phosphorylated forms, whereas p107 is not detectable (Lin and DeCaprio 2003). In mid-G1/S transition, RB family proteins are phosphorylated by cyclin/CDK complexes. Hypo-ppRB and non- and low-phosphorylated p130 change to hyper-ppRB and high-phosphorylated p130, respectively. p107 is synthesized de novo, followed by phosphorylation to a high-phosphorylated form. Protein levels of pRB remain relatively constant throughout the cell cycle, compared to either p130 or p107. Although there is abundant p130 in the G0/G1 phase, it is dramatically down-regulated in the mid-G1/S phase. p130 down-regulation appears to be mediated by a cell cycle stage-dependent proteasome degradation pathway (Tedesco et al. 2002). These observations suggest that RB family proteins have overlapping functions, as well as different functions in the cell cycle regulation of quiescent cells.
In plants, knowledge of cell cycle control has increased in recent years (Gegas and Doonan 2006). Some basic mechanisms that regulate the cell cycle appear to have been conserved throughout eukaryotic evolution (Dewitte and Murray 2003). To date, plant cDNAs encoding RB-related proteins have been reported in Zea mays (Ach et al. 1997; Grafi et al. 1996; Xie et al. 1996), Nicotiana tabacum (Nakagami et al. 1999), Chenopodium rubrum (Fountain et al. 1999), Arabidopsis thaliana (Kong et al. 2000), Populus tremula × Populus tremuloides (AF133675), Cocos nucifera (AY117036), and Oryza sativa (AP004592). The amino acid conservation between animal and plant RB-related proteins suggests that the proteins have similar biochemical properties. Like animal RB family proteins, plant RB-related proteins also interact with various cellular proteins, such as E2F family proteins, D-type cyclin, mammalian viral oncoproteins (SV40 large-T antigen, adenovirus E1a, and HPV E7), and the plant virus proteins (wheat dwarf virus RepA, and the tomato golden mosaic virus replication factor AL1) (Ach et al. 1997; Grafi et al. 1996; Huntley et al. 1998; Nakagami et al. 1999; Ramirez-Parra et al. 1999; Sekine et al. 1999; Xie et al. 1996).
Because the functions of the mammalian RB family proteins depend on their phosphorylation state, the phosphorylation states of plant RB-related proteins were analyzed. ZmRBR1 protein undergoes changes in phosphorylation states concomitant with endoreduplication in maize (Grafi et al. 1996). The human G1/S protein kinases cyclinD/CDK4, cyclinE/CDK2, and cyclinA/CDK2 can phosphorylate ZmRBR1 protein in vitro (Huntley et al. 1998), and NtRBR1 protein is phosphorylated by tobacco cyclinD/CDC2 in vitro (Nakagami et al. 1999). The ZmRBR1 kinase activity correlates with the proliferation state in maize leaf (Boniotti and Gutierrez 2001), and NtRBR1 kinase activity is detected only during the mid-G1/S phase in tobacco BY-2 cells (Nakagami et al. 2002). These observations also suggest that plant RB-related proteins have biochemical properties similar to those of mammalian RB family proteins. Plant RB-related proteins seem to control not only cell cycle arrest/progression, but also development and cellular differentiation in endosperm, leaf, and root (Ebel et al. 2004; Desvoyes et al. 2006; Wildwater et al. 2005). Only limited information is available, however, on the development-dependent phosphorylation states of plant RB-related protein. In this paper, we describe the isolation and functional characterization of a pea (Pisum sativum L. cv. Alaska) cDNA encoding an RB-related protein (PsRBR1), which has biochemical properties similar to those of mammalian RB family proteins. PsRBR1 protein undergoes changes in its phosphorylation state concomitant with dormancy-to-growth transition in pea axillary buds.
Materials and methods
Plant growth and tissue collection
Seeds of Pisum sativum L. cv. Alaska were soaked in running tap water for 24 h and sown in trays of rockwool. Plants were grown at 25°C in the dark for 4 days, and then in a 16 h light/8 h dark photoperiod for 3 days. Tissues studied were axillary buds at the second node in 7-day-old seedlings. Plants were decapitated 1 cm above the second node to stimulate outgrowth of axillary buds.
PCR and cloning of PsRBR1 cDNA
Degenerate oligonucleotide primers were designed for conserved regions of published amino acid sequences of the RB family proteins (sense; 5′-TT(T/C)TT(T/C)AA(T/C)C GNCA(T/C)AT(T/C/A)GA(T/C)CA-3′ and antisense; 5′-AC(T/C)TC(G/A)TT(G/A)TA(G/A)AANGT(T/G/A)AT(T/G/A)AT-3′), where the N in the parentheses indicates all four deoxyribonucleotides. Polymerase chain reaction (PCR) amplification was performed with cDNA from total RNAs of shoot apices. The amplified fragments (237 bp) were cloned into a BSII TSK-plasmid vector (Ichihara and Kurosawa 1993) and sequenced. A pea cDNA library was constructed using poly (A)+ RNA prepared from dormant axillary buds with a HybriZAP-2.1 A Two-Hybrid cDNA Gigapack Cloning Kit (Stratagene, La Jolla, CA). Approximately 1 × 106 phage recombinants derived from the pea cDNA library were screened with the selected PCR products labeled with [32P]-dCTP. Hybridization and subsequent washing were performed as described by Shimizu and Mori (1998).
DNA sequencing
The cDNAs were sequenced by the dideoxy chain termination method using an automatic DNA sequencer (LIC-4000, LI-COR Inc., Lincoln, NE) according to the manufacturer’s protocol. Both strands were entirely sequenced.
Yeast two-hybrid system
The vectors and strains were provided in the MATCHMAKER Two-Hybrid System 2 (Clontech, Palo Alto, CA). DNA manipulations and yeast methods were performed according to the manufacturer’s instructions. PsRBR1 cDNA was introduced into the pAS2-1 vector, and another cDNA was introduced into the pACT2 vector. Saccharomyces cerevisiae Y187 was transformed with combinations of these plasmids. For quantitative liquid assay, β-galactosidase activity was assayed with O-nitrophenylgalactoside as a substrate.
Antibody preparation and immunoblot analysis
For an anti-PsRBR1 antibody, full-length PsRBR1 protein was expressed in Escherichia coli BL21 pLysS as a 6xHis-tagged protein in pET32a (Novagen, Madison, WI). The antigen was purified with TALON Metal Affinity Resin (Clontech) according to the manufacturer’s protocol. The antigen was injected into a mouse using conventional procedures.
Proteins from axillary buds were extracted by grinding with aluminum oxide in extraction buffer (20 mM Tris–HCl [pH 8.0], 100 mM NaCl, 1 mM EDTA, 0.5% NP-40, 50 μM NaF, and 1 μM Na3VO4, and the proteinase inhibitor cocktail Complete [Roche, Penzberg, Germany]). Insoluble materials were removed by microcentrifugation. Protein concentrations were determined with the DC Protein Assay (Bio-Rad, Hercules, CA). Proteins were separated using sodium dodecyl sulfate—polyacrylamide gel electrophoresis (SDS-PAGE, 10% acrylamide gels [acrylamide:bis-acrylamide = 37.5:1]), or 10% low bis-acrylamide gels [acrylamide:bis-acrylamide = 142:1]), and blotted onto a nitrocellulose membrane (BA-S 85, Schleicher & Schuell, Dassel, Germany). The membrane was blocked with 5% skim milk and 0.05% Tween 20 in Tris-buffered saline (50 mM Tris–HCl [pH 8.0], and 150 mM NaCl), and incubated with the anti-PsRBR1 antibody at a dilution of 1:1000. Horseradish peroxidase-conjugated goat anti-mouse IgG (Pierce, Rockford, IL) was used as the secondary antibody at a dilution of 1:2000. The signals were detected with the SuperSignal ULTRA (Pierce).
In vitro-coupled transcription and translation
A cDNA fragment corresponding to PsRBR1 cDNA, encoding the full-length protein, was cloned into the pBluescript SK+ vector (Stratagene). The template was digested with a restriction enzyme for linearization. In vitro-coupled transcription-translation was performed with the digested template (1 μg) in a TnT® T3 Coupled Wheat Germ Extract System (Promega, Madison, WI) in the presence of [35S]-methionine according to the manufacturer’s protocol. As a control, the in vitro-coupled transcription-translation reaction was performed without the DNA template. Samples were separated by SDS-PAGE. The gel was dried and exposed to a Fuji Imaging Plate (Fuji Photo Film, Tokyo, Japan) overnight. The radiographic image was visualized with a BAS2000 Image Analyzer (Fuji Photo Film).
Dephosphorylation
Protein extracts were incubated in reaction mixture (50 mM Hepes [pH 7.5], 2 mM MnCl2, 0.1 mM EDTA, 5 mM dithiothreitol, 0.01% Brij35, and the proteinase inhibitor cocktail Complete) with 1 unit of lambda protein phosphatase (λ-PPase, New England Biolabs, Beverly, MA) for 30 min at 30°C. Samples were immunoblotted by the anti-PsRBR1 antibody as described above.
RNA extraction and blot analysis
Total RNA was isolated from the tissues by SDS-phenol extraction, followed by LiCl precipitation. Formaldehyde agarose gel electrophoresis of total RNA was performed using standard procedures. The RNAs were blotted onto a Hybond N+ membrane (Amersham Biosciences), and hybridized with PsRBR1, PCNA, and Histone H4 cDNA labeled with [32P]-dCTP, respectively. Hybridization was performed in 0.25 M NaH2PO4 buffer (pH 7.2), 0.25 M NaCl, 7% SDS, 10% dextran sulfate (Amersham Biosciences), and 1% polyvinylpyrrolidone K30 at 65°C for at least 16 h. The membrane was washed in 2xSSPE (1xSSPE is 0.15 M NaCl, 10 mM NaH2PO4, and 1 mM EDTA [pH 7.4]) and 0.1% SDS at 50°C twice for 15 min each, and visualized by autoradiography at −80°C using Kodak XAR-5 film and an intensifying screen (Kodak, Rochester, NY).
Phospholabeling and immunoprecipitation
[32P]-inorganic phosphate (Amersham Biosciences, Piscataway, NJ) was applied to axillary buds (10 μCi per bud) 1 h before collection of the buds. Protein extraction was performed as described above. Protein extracts were incubated with both preimmune serum and Protein A Sepharose 4 Fast Flow (Amersham Biosciences) for 1.5 h at 4°C, and then non-specific associated materials were removed by microcentrifugation. The anti-PsRBR1 antibody was added to the clarified extracts and incubated for 1 h on ice. Protein A Sepharose 4 Fast Flow was added, and incubated for 30 min at 4°C. The complex of immunoprecipitates was washed three times with extraction buffer with 1 mg/ml bovine serum albumin, resuspended in SDS sample buffer (20 mM Tris-HCl [pH 6.8], 40% glycerol, 2% SDS, 2% β-mercaptoethanol, and bromophenol blue), and separated by SDS-PAGE. Dried gels were placed on a Fuji Imaging Plate, and the radiographic images were analyzed using the BAS2000 Image Analyzer.
Results
Isolation of a pea cDNA encoding RB-related protein
To analyze the molecular mechanisms of cell cycle control in axillary buds, we focused on the function of plant RB-related proteins during dormancy-to-growth transition. Based on the amino acid sequences of reported RB-related proteins, we designed degenerate primers and performed reverse transcription PCR with total RNA prepared from pea shoot apices. Using the amplified DNA fragments as a probe, we screened the cDNA library prepared from pea dormant axillary buds. We obtained 14 positive clones by screening, and partially sequenced their 5′ ends. The clone that had the longest 5′ sequence (3540 bp) was used for further analysis because the DNA sequences of the other clones corresponded to those of the longest clone. This cDNA contained a single open reading frame capable of encoding a 1026-amino acid residue protein, PsRBR1. The molecular mass of PsRBR1 protein was estimated to be 114-kDa. The PsRBR1 cDNA contained a stop codon (TAG) four amino acids upstream of the putative initiation methionine codon within the reading frame, indicating that the PsRBR1 cDNA contained the entire coding region.
The alignment of the A and B conserved domains and the N-terminal leucine-rich domain of PsRBR1 protein with maize ZmRBR1 protein and human RB family proteins is shown in Fig. 1(A). The A and B conserved domains of the PsRBR1 protein share 43% identity and 71% similarity, respectively with the A and B conserved domains of the ZmRBR1 protein. The conserved cysteine residue corresponding to Cys 706 of human pRB was present as Cys 798 in PsRBR1 protein (shown by an asterisk in the B conserved domain). This conserved cysteine residue is essential for the function of mammalian and plant RB proteins (Ach et al. 1997; Grafi et al. 1996; Kaye et al. 1990).
Comparison of deduced amino acid sequences of pea PsRBR1 with selected homologous sequences. (A) Sequence alignments of the A and B conserved domains, and the N-terminal leucine-rich domain of PsRBR1 protein and ZmRBR1 protein with human RB family proteins. Black boxes indicate conserved amino acid residues, and dashes indicate gaps in the sequences. The asterisk in the B conserved domain denotes a conserved cysteine residue (798 of PsRBR1 protein), essential for RB function in mammalian cells. (B) Phylogenic analysis of the RB-related proteins under neighbor-joining analysis, and schematic of the salient features of RB-related proteins. The phylogeny is based on the amino acid sequences from the A conserved domain of RB-related proteins. Branch lengths are indicated above each branch. Bootstrap values from the results of 1000 replicates are shown below a node if the node is present in more than 50% of the bootstrap replicate analyses. The three conserved regions (N, N-terminal leucine-rich domain; A, A conserved domain; B, B conserved domain) between plants and mammals are indicated in boxes. Bars represent potential CDK phosphorylation sites. Numbers refer to protein length in amino acids, and numbers in parentheses show potential CDK phosphorylation sites. The accession numbers are AtRBR1 (AC069472), CnRBR1 (AY117036), CrRBR1 (AJ011681), NtRBR1 (AB015221), OsRBR1 (AP004592), PsRBR1 (AB012024), PtRBR1 (AF133675), ZmRBR1 (AF250050), p107 (L14812), p130 (X76061), and pRB (AF551763)
The phylogenic analysis of plant RB-related proteins reported so far and human RB family proteins is shown in Fig. 1(B). The scheme on the right in Fig. 1(B) shows a diagram of the characteristic features of RB-related proteins. PsRBR1 protein was most similar to Populus tremula × Populus tremuloides PtRBR1 protein. PsRBR1 protein has 16 potential phosphorylation sites (Ser-Pro or Thr-Pro) for CDKs, which are highly clustered in regions flanking the A and B conserved domains. These conserved primary structures strongly suggest that RB-related proteins, including PsRBR1 protein, have similar biochemical properties in plant and animals.
Genomic Southern blot analyses were performed to estimate the copy number and closely related genes of PsRBR1 in pea. Full-length PsRBR1 cDNA was used as a probe. Hybridization at both high- and low-stringencies yielded only signals derived from PsRBR1 (data not shown). These results suggested that PsRBR1 gene exists as a single-copy gene in pea.
Interactions of PsRBR1 protein with both plant D-type cyclins and a mammalian oncoprotein
Mammalian and plant RB proteins bind to a large number of cellular and viral proteins, such as D-type cyclin and SV40 large-T antigen. To confirm that PsRBR1 protein acts in the same manner as the mammalian and various plant RB proteins, we tested the ability of PsRBR1 protein to interact with these proteins using a yeast two-hybrid system. The genes for the tested proteins were fused to the GAL4 DNA-binding domain or the GAL4 activation domain. The data shown in Table 1 are the averages of three independent transformants. We verified that PsRBR1 protein alone did not activate the reporter gene. PsRBR1 protein interacted with both pea and Arabidopsis D-type cyclin (Pissa;CycD3;1 and Arath;CycD3;1). Both D-type cyclins contain the canonical RB-binding motif, LXCXE, in their N-terminus (Shimizu and Mori 1998; Soni et al. 1995). PsRBR1 protein interacted with SV40 large-T antigen, which is a mammalian DNA tumor virus oncoprotein. The similarities in primary structures and the coincidence of interacting partners of RB-related proteins suggest that animal and plant RB-related proteins, including PsRBR1 protein, have similar functions in cell cycle regulation.
Immunochemical detection of PsRBR1 protein in pea axillary buds
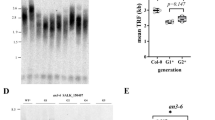
To investigate protein levels and phosphorylation states of PsRBR1 protein in axillary buds during dormancy-to-growth transition, we prepared an anti-PsRBR1 antibody. The specificity of the anti-PsRBR1 antibody against PsRBR1 protein was examined (Fig. 2A). Protein extracted from growing axillary buds was detected by immunoblotting with the anti-PsRBR1 antibody. The anti-PsRBR1 antibody cross-reacted with three polypeptides (Fig. 2A, lane 1, filled circle, empty circle, and arrowhead), with molecular masses of approximately 150, 140, and 114-kDa. The pre-immune serum did not cross-react with any polypeptides in the protein extracts (Fig. 2A, lane 2). The anti-PsRBR1 antibody was incubated with an excess of purified recombinant PsRBR1 protein as a competitor, and then used to detect PsRBR1 protein in pea extract (Fig. 2A, lane 3). In this case, no polypeptides were detected with the anti-PsRBR1 antibody. These results indicate that the anti-PsRBR1 antibody specifically cross-reacts with PsRBR1 protein in axillary bud extracts.
Immunochemical detection of PsRBR1 protein in pea axillary buds. (A) The terminal bud was cut 1 cm above the second node. After 24 h, the growing axillary buds were collected. Proteins in the protein extracts (30 μg) prepared from axillary buds were detected by immunoblotting with the anti-PsRBR1 antibody (lane 1), pre-immune serum (lane 2), and anti-PsRBR1 antibody preincubated with excess recombinant PsRBR1 protein (lane 3). Molecular mass markers are indicated at the left of the lane. The anti-PsRBR1 antibody cross-reacted with three polypeptides, indicated by the filled circle, empty circle, and arrowhead. (B) Comparison of molecular mass of authentic PsRBR1 protein with that of PsRBR1 synthesized by in vitro-coupled transcription-translation reaction. The plasmid DNA containing the T3 promoter and the PsRBR1 coding sequence was transcribed, and the resultant RNA was translated. The translation products labeled with [35S]-methionine (lane 2) were immunoprecipitated by the anti-PsRBR1 antibody (lane 3). Proteins in the protein extracts prepared from growing axillary buds were detected by immunoblotting with the anti-PsRBR1 antibody (lane 4). The in vitro-coupled transcription-translation reaction was performed without the plasmid DNA (lane 1). Molecular mass markers are indicated at the left of the lane. The product synthesized in vitro was a 114-kDa polypeptide, indicated by the arrowhead (lane 2), which is consistent with the calculated values based on the deduced amino acids sequences of PsRBR1 cDNA. (C) Dephosphorylation of authentic PsRBR1 protein. Protein extracts prepared from growing axillary buds were dephosphorylated with λ-PPase treatment, separated by SDS-PAGE in either a standard acrylamide gel (left) or low bis-acrylamide gel (right), and proteins were detected by immunoblotting with the anti-PsRBR1 antibody. Numbers in parentheses indicate the ratio of acrylamide to bis-acrylamide. Lane 1, untreated samples; lane 2, samples incubated without λ-PPase; lane 3, samples incubated with λ-PPase
Comparison of molecular mass of authentic PsRBR1 protein in pea with that of in vitro-synthesized PsRBR1 protein
Three polypeptides were detected in protein extracts from pea axillary buds by immunoblotting with the anti-PsRBR1 antibody. The molecular mass of PsRBR1 protein was estimated to be 114-kDa based on the deduced amino acid sequences of PsRBR1 cDNA. To clarify these differences in the molecular mass of PsRBR1 protein, we compared the molecular mass of authentic PsRBR1 protein in pea with that of in vitro-synthesized PsRBR1 protein (Fig. 2B). PsRBR1 protein was synthesized using an in vitro-coupled transcription-translation system with [35S]-methionine. In vitro-synthesized PsRBR1 protein produced a major 114-kDa polypeptide (Fig. 2B, lane 2, arrowhead). The molecular mass of the PsRBR1 protein was consistent with that predicted from the deduced amino acid sequences of PsRBR1 cDNA. The 114-kDa polypeptide was not detected in protein products prepared from the in vitro-coupled transcription-translation system without the PsRBR1 DNA template, which was used as a negative control (Fig. 2B, lane 1). To confirm whether the 114-kDa polypeptide was PsRBR1 protein, in vitro-coupled transcription-translation products were immunoprecipitated with the anti-PsRBR1 antibody (Fig. 2B, lane 3). The 114-kDa polypeptide was immunoprecipitated with the anti-PsRBR1 antibody, indicating that in vitro-synthesized PsRBR1 protein is 114-kDa and corresponds to the smallest polypeptide detected by anti-PsRBR1 antibody in growing axillary buds (Fig. 2B, lane 4). These results suggested that the smallest polypeptide (114-kDa) detected by the anti-PsRBR1 antibody was non-modified PsRBR1 protein in axillary buds, and the two larger polypeptides of PsRBR1 proteins were modified by post-translational regulation in axillary buds.
Dephosphorylation of authentic PsRBR1 protein
The activity of mammalian RB family proteins is regulated by their phosphorylation states. Plant RB-related proteins are also phosphorylated by cyclin/CDK complexes (Huntley et al. 1998; Nakagami et al. 1999). PsRBR1 protein has some putative CDK phosphorylation sites (Fig. 1B). Thus, we expected that the modification of authentic PsRBR1 protein in the axillary buds caused its phosphorylation. To examine this, we performed in vitro dephosphorylation reactions against PsRBR1 protein using (λ-PPase. Proteins were separated by two types of SDS-PAGE gels, because some phosphorylated proteins were more clearly separated by low bis-acrylamide gel (Fig. 2C, right) than by standard acrylamide gel (Fig. 2C, left) (Iwasaki et al. 2002). Immunoblotting with anti-PsRBR1 antibody indicated that there were three polypeptides corresponding to PsRBR1 protein in growing axillary buds (Fig. 2C, lane 1), but after λ-PPase treatment of the protein extract, PsRBR1 protein was detected as the smallest of the polypeptides (Fig. 2C, lane 3). On the other hand, the molecular mass of PsRBR1 protein did not change after treatment without λ-PPase added to the protein extract as a negative control (Fig. 2C, lane 2). In addition, the low bis-acrylamide gel separated the protein more clearly than did the standard acrylamide gel. These results suggested that PsRBR1 protein was controlled by post-translational modification, and exists in non-phosphorylated form of PsRBR1 protein, lower-molecular mass form of phosphorylated PsRBR1 protein (Lp-form), and higher-molecular mass form of phosphorylated PsRBR1 protein (Hp-form) in growing axillary buds.
Protein levels and phosphorylation states of PsRBR1 protein in axillary buds during dormancy-to-growth transition
Because suppression of the cell cycle is regulated by the phosphorylation states of mammalian RB family proteins, we evaluated the phosphorylation states of PsRBR1 during dormancy-to-growth transition in pea axillary buds by immunoblotting using the anti-PsRBR1 antibody (Fig. 3A). During dormancy, PsRBR1 protein was detected as two polypeptides, which are non-phosphorylated form of PsRBR1 protein and the Lp-form (Fig. 3A, 0 h after decapitation). In addition to these non-phosphorylated form of PsRBR1 protein and the Lp-form, the Hp-form was detected 2 h after decapitation (Fig. 3A, indicated by the arrow). These results suggested that PsRBR1 protein was shifted from non-phosphorylated form of PsRBR1 protein and the Lp-form to the Hp-form; that is, the PsRBR1 protein was more phosphorylated after decapitation. The protein levels of these three polypeptides were increased 12 h after decapitation. Non-phosphorylated PsRBR1 protein and the Lp-form existed during late stage (12 to 24 h) after decapitation. PsRBR1 mRNA level was increased 12 h after decapitation (see Fig. 3B). It seems that non-phosphorylated PsRBR1 protein and the Lp-form were synthesized and phosphorylated de novo after decapitation. Moreover, because the axillary bud cell cycle is not completely synchronized, the non-phosphorylated PsRBR1 protein and the Lp-form were included in the protein extracts prepared from growing axillary buds.
Protein levels and phosphorylation states of PsRBR1 protein in axillary buds during the dormancy-to-growth transition. (A) Proteins were extracted from axillary buds after terminal bud decapitation. Numbers below each lane indicate the hours after decapitation. Proteins in the protein extracts (30 μg) were detected by immunoblotting with the anti-PsRBR1 antibody. Molecular mass markers are indicated at the left of the lane. The bottom panel shows the Coomassie Brilliant Blue (CBB) staining around 100 kDa as a loading control. The filled circle, empty circle, and arrowhead indicate higher-molecular mass form of phosphorylated PsRBR1 protein (Hp-form), lower-molecular mass form of phosphorylated PsRBR1 protein (Lp-form) and non-phosphorylated PsRBR1 protein, respectively. The arrow shows more phosphorylated PsRBR1 protein (Hp-form) appearing 2 h after decapitation. (B) Northern blot analyses of total RNA isolated from pea axillary buds before and after decapitation. Plants were decapitated 1 cm above the second node. After decapitation, axillary buds were collected at several time points, as indicated below each lane. Total RNA (10 μg) was separated by denaturing formaldehyde gel electrophoresis, and transferred to a Hybond N+ membrane. The membranes were hybridized with the cDNA of PsRBR1, PCNA, and histone H4. The bottom panel shows the ethidium bromide-stained RNA gel as a loading control
Accumulation patterns of PsRBR1 transcripts in axillary buds during dormancy-to-growth transition
To investigate the relationship between different phosphorylation states of PsRBR1 protein with the transcriptional control of cell cycle regulators, we examined the accumulation patterns of PsRBR1, PCNA, and Histone H4 transcripts during dormancy-to-growth transition in axillary buds. The results of Northern blot analyses using PsRBR1, PCNA, and histone H4 cDNAs (Shimizu and Mori 1998) are shown in Fig. 3B. PsRBR1 transcripts were detected at 3.6 kb. The mRNA levels of the three genes were very low during dormancy in axillary buds. When the axillary buds were stimulated to grow by terminal bud decapitation, mRNA levels of these genes increased markedly. PsRBR1 and histone H4 mRNA increased 12 h after decapitation. Histone H4 was specifically expressed during the S phase. The accumulation patterns of PsRBR1 mRNA are similar to that of histone H4 mRNA, suggesting that PsRBR1 is predominantly expressed at the S phase in pea axillary buds at least during the first 24 h after terminal bud decapitation. Because PsRBR1 protein was increased 12 h after decapitation (see Fig. 3A), the expression patterns of PsRBR1 are coincident with those of PsRBR1 protein. The mRNA expression of PCNA seems to be regulated by RB-related proteins. PCNA mRNA increased significantly 4 h after decapitation, and PCNA mRNA accumulated after the appearance of the Hp-form.
Detection of phosphorylated PsRBR1 protein in axillary buds after decapitation
We confirmed that PsRBR1 protein was phosphorylated in the axillary buds just after decapitation with in vivo labeling followed by immunoprecipitation using the anti-PsRBR1 antibody (Fig. 4). [32P]-inorganic phosphate was incorporated into growing axillary buds. Many proteins were phosphorylated in growing axillary buds after decapitation (Fig. 4A, lane 1). Immunoprecipitation with the anti-PsRBR1 antibody revealed a [32P]-labeled 150-kDa phosphoprotein (Fig. 4A, lane 3, arrow). The size of the [32P]-labeled PsRBR1 protein was consistent with the Hp-form detected by immunoblotting analysis. The signal was not detected by immunoprecipitation with preimmune serum used as a negative control (Fig. 4A, lane 2).
Detection of phosphorylation of PsRBR1 protein in axillary buds during dormancy-to-growth transition. (A) Eight hours after decapitation of the terminal bud, [32P]-inorganic phosphate was applied to axillary buds to be incorporated in vivo. One hour after the application of [32P]-inorganic phosphate, proteins were extracted from the axillary buds. The protein extracts were immunoprecipitated with the anti-PsRBR1 antibody (lane 3) or preimmune serum (lane 2), and separated by SDS-PAGE. Total proteins without immunoprecipitation were loaded in lane 1. The dried gel was exposed to a Fuji Imaging Plate. Molecular mass markers are indicated on the left of the lane. (B) [32P]-inorganic phosphate was applied to axillary buds 1 h before collection of the buds. The axillary buds were collected at 0, 2, and 4 h after terminal bud decapitation (lanes 1 to 3, respectively), and proteins were extracted from the axillary buds. Other details are as described in (A)
To confirm whether PsRBR1 protein is more phosphorylated in axillary buds just after decapitation, we examined dormant and outgrowing axillary buds after decapitation. No signal was detected before decapitation (Fig. 4B, lane 1), suggesting that PsRBR1 protein was not highly phosphorylated de novo before decapitation. Two hours after decapitation, the 150-kDa polypeptide was detected (Fig. 4B, lane 2) and the signal was increased 4 h after decapitation (Fig. 4B, lane 3, arrow). These results suggested that the level of phosphorylation of PsRBR1 protein increased immediately after terminal bud decapitation, producing the Hp-form.
Discussion
In intact plants, the pea shoot apex inhibits growth; i.e., cell elongation and cell division of the dormant axillary buds. The cell cycle of dormant axillary buds is arrested at the G1 phase. Therefore, from the point of view of cell cycle control, the transition from dormancy to outgrowth in axillary buds seems to be from the G1 phase to the S phase. In animal cells, RB family proteins function as negative controllers in G1 arrest, and the activities of RB family proteins are controlled by their phosphorylation states. Here, we investigated phosphorylation states of the pea RB-related protein, PsRBR1, during the dormancy-to-growth transition in axillary buds. The results of the present study provide a basis for further studies of the molecular mechanisms underlying apical dominance and cell cycle control.
Mammalian and plant RB proteins have several highly conserved regions, which are the N-terminal leucine rich, A and B conserved domains, multiple potential CDK phosphorylation sites, and a conserved cysteine residue. PsRBR1 protein, like other plant RB-related proteins, has the ability to interact with both plant D-type cyclins and a mammalian oncoprotein. Plant RB-related proteins have the same properties and activities as animal RB family proteins. For example, ZmRBR1 protein binds to human and Drosophila E2F, and inhibits transcriptional activation of human E2F (Huntley et al. 1998; Ramirez-Parra et al. 1999; Sekine et al. 1999). Geminivirus DNA replication is reduced in plant cells transfected with plasmids encoding human p130 (Xie et al. 1996). Human cyclinD/CDKA phosphorylates NtRBR1 protein in vitro (Nakagami et al. 1999). Thus, plant RB-related proteins can function not only as plant factors, but also as animal factors. These results strongly suggest that the functions of plant RB-related proteins, including PsRBR1 protein, are similar to those of the mammalian RB family proteins in G1/S transition, and that plant and animal cells might use similar regulatory proteins and pathways for cell cycle control during the G1 phase. Therefore, the maintenance of this high degree of similarity throughout evolution (in plants, insects, and vertebrates), further emphasizes the critical roles of RB-related proteins.
Three human RB family proteins (pRB, p130, and p107) have individual functions in cell cycle regulation and development. Two RB-related genes were isolated from Drosophila melanogaster. To date, plant RB-related proteins have been reported in Zea mays, Nicotiana tabacum, Chenopodium rubrum, Arabidopsis thaliana, Populus tremula x Populus tremuloides, Cocos nunucifera, and Oryza sativa (Ach et al. 1997; Fountain et al. 1999; Grafi et al. 1996; Kong et al. 2000; Nakagami et al. 1999; Xie et al. 1996). Of these, Arabidopsis thaliana and Oryza sativa might contain only one copy of the RB-related gene in the whole genome based on the similarity of amino acid sequences of RB-related proteins. Here, our genomic Southern blot analysis suggested the presence of only one copy of the PsRBR1 gene in pea. The dicot RB-related gene seems to contain only one copy in the genome. By contrast, maize contains at least three RB-related genes. ZmRBR3 protein also functions to promote endosperm development (Sabelli et al. 2005), suggesting that maize RB family proteins have special roles in endosperm development.
When the protein extracts in growing axillary buds were treated with λ-PPase, the Lp- and Hp-forms should have been dephopshorylated and stoichiometrically shifted to non-phosphorylated forms. As shown in Fig. 3(C), however, it appears that there was no increase in the non-phosphorylated form (lane 3) compared to the other lanes. The reason for this might be that a 40-kDa polypeptide was also detected by immunoblotting with the anti-PsRBR1 antibody (data not shown). The 40-kDa polypeptide was probably derived from in vitro-dephosphorylated PsRBR1 protein by degradation during λ-PPase treatment. In vitro-dephosphorylated PsRBR1 protein might be more unstable than in vivo-dephosphorylated PsRBR1 protein, despite the presence of proteinase inhibitors, though the reason for this is not clear.
The results of the immunoblotting experiments indicated that the migration patterns of mammalian RB family proteins differ depending on their phosphorylation states. According to the reports (Farkas et al. 2002), the faster migrating proteins were the non- and hypo-phosphorylated forms in the G0/G1 phases. The slower migrating protein was the hyper-phosphorylated form during the G1/S transition. In plant, immunoblotting with the anti-ZmRBR1 antibody revealed the ZmRBR1 protein to be a diffuse protein band of approximately 110-kDa during endoreduplication in maize endosperm, and there are some polypeptides of approximately 110-kDa in maize leaf (Grafi et al. 1996; Huntley et al. 1998). The diffused protein bands of ZmRBR1 were confirmed by the use of λ-phosphatase to result from phosphorylation. (Grafi et al. 1996). In the present paper, using λ-PPase treatment and in vivo labeling followed by immunoprecipitation with the anti-PsRBR1 antibody, we showed that PsRBR1 protein exists in three phosphorylated forms in axillary buds. Although we could not determine the number of phosphorylation sites in the Lp- and Hp-forms, the Lp- and Hp-forms might correspond to the hypo- and hyper-phosphorylated forms of PsRBR1 protein, respectively.
Plant E2F isolated from tobacco and wheat has transcriptional activity and interacts with RB-related protein (Ramirez-Parra et al. 1999; Sekine et al. 1999). There are E2F-binding sites in the promoter region of PCNA, whose transcripts are induced in the late-G1/S phase (Kodama et al. 1991). These observations suggest that plant E2F, like that in animal, induces gene expression of PCNA. Further phosphorylation of PsRBR1 protein was induced 2 h after terminal bud decapitation, and PCNA transcripts were induced 4 h after terminal bud decapitation in pea axillary buds. Although it is unclear whether the transcriptional activation of plant PCNA is regulated by E2F, E2F released from RB-related protein might promote PCNA mRNA expression, as in mammals. This possibility is supported by the observation that ZmRBR1-associated kinase activity peaks at the G1/S transition in synchronized tobacco BY-2 cells (Boniotti and Gutierrez 2001). It is likely that increased phosphorylation states of PsRBR1 protein have an important role in the regulation of E2F-mediated gene expression involved in the G1/S transition and DNA replication during dormancy-to-growth transition in pea axillary buds.
In some plants, the gene expression of cyclins and CDK activities are regulated by plant hormones, e.g., cytokinin, auxin, and gibberellin (Miao et al. 1993; Sauter 1997; Soni et al. 1995). In Arabidopsis suspension-cultured cells, D-type cyclin mRNA is rapidly induced by the addition of cytokinin or glucose, and repressed by the addition of auxin (Soni et al. 1995). Cytokinin is biosynthesized at the stem after decapitation and moves into the axillary buds for outgrowth (Tanaka et al. 2006). It is possible that the increasing cytokinin levels caused by decapitation induces the expression of D-type cyclin. D-type cyclin forms a complex with CDKs and immediately phosphorylates RB-related protein. The Hp-form induces gene expression for the G1/S transition in axillary bud cells. It is likely that RB-related protein controls the cell cycle in axillary buds during the dormancy-to-growth transition. Further analyses will provide more insight into the regulation of dormancy-to-growth transition in axillary buds.
Abbreviations
- CDK:
-
cyclin-dependent kinase
- Hp-form:
-
higher-molecular mass form of phosphorylated PsRBR1 protein
- λ-PPase:
-
lambda protein phosphatase
- Lp-form:
-
lower-molecular mass form of phosphorylated PsRBR1 protein
- PCNA:
-
proliferating cell nuclear antigen
- PCR:
-
polymerase chain reaction
- pRB:
-
phosphoprotein retinoblastoma tumor suppressor
- ppRB:
-
phosphorylated pRB
- RB :
-
retinoblastoma tumor suppressor gene
- SDS-PAGE:
-
sodium dodecyl sulfate—polyacrylamide gel electrophoresis
References
Ach RA, Durfee T, Miller AB, Taranto P, Hanley-Bowdoin L, Zambryski PC, Gruissem W (1997) RRB1 and RRB2 encode maize retinoblastoma-related proteins that interact with a plant D-type cyclin and geminivirus replication protein. Mol Cell Biol 17:5077–5086
Bennett T, Leyser O (2006) Something on the side: axillary meristems and plant development. Plant Mol Biol 60:843–854
Boniotti MB, Gutierrez C (2001) A cell-cycle-regulated kinase activity phosphorylates plant retinoblastoma protein and contains, in Arabidopsis, a CDKA/cyclin D complex. Plant J 28:341–350
Booker J, Chatfield S, Leyser O (2003) Auxin acts in xylem-associated or medullary cells to mediate apical dominance. Plant Cell 15:495–507
Desvoyes B, Ramirez-Parra E, Xie Q, Chua N-H, Gutierrez C (2006) Cell type-specific role of the retinoblastoma/E2F pathway during Arabidopsis leaf development. Plant Physiol 140:67–80
Devitt ML, Stafstrom JP (1995) Cell cycle regulation during growth-dormancy cycles in pea axillary buds. Plant Mol Biol 29:255–265
Dewitte W, Murray JAH (2003) The plant cell cycle. Annu Rev Plant Biol 54:235–264
Dun EA, Ferguson BJ, Beveridge CA (2006) Apical dominance and shoot branching. Divergent opinions or divergent mechanisms? Plant Physiol 142:812–819
Ebel C, Mariconti L, Gruissem W (2004) Plant retinoblastoma homologues control nuclear proliferation in the female gametophyte. Nature 429:776–780
Farkas T, Hansen K, Holm K, Lukas J, Bartek J (2002) Distinct phosphorylation events regulate p130- and p107-mediated repression of E2F-4. J Biol Chem 277:26741–26752
Fountain MD, Murray JAH, Beck E (1999) Isolation of a full-length cDNA encoding a retinoblastoma protein (Accession No. AJ011681) from suspension-cultured photoautotrophic Chenopodium rubrum L. cells. Plant Physiol 119:363
Gegas VC, Doonan JH (2006) Expression of cell cycle genes in shoot apical meristems. Plant Mol Biol 60:947–961
Grafi G, Burnett RJ, Helentjaris T, Larkins BA, DeCaprio JA, Sellers WR, Kaelin WG Jr (1996) A maize cDNA encoding a member of the retinoblastoma protein family: involvement in endoreduplication. Proc Natl Acad Sci USA 93:8962–8967
Graña X, Garriga J, Mayol X (1998) Role of the retinoblastoma protein family, pRB, p107 and p130 in the negative control of cell growth. Oncogene 17:3365–3383
Huntley R, Healy S, Freeman D, Lavender P, de Jager S, Geenwood J, Makker J, Walker E, Jackman M, Xie Q, Bannister AJ, Kouzarides T, Gutierrez C, Doonan JH, Murray JAH (1998) The maize retinoblastoma protein homologue ZmRb-1 is regulated during leaf development and displays conserved interactions with G1/S regulators and plant cyclin D (CycD) proteins. Plant Mol Biol 37:155–169
Ichihara Y, Kurosawa Y (1993) Construction of new T vectors for direct cloning of PCR products. Gene 130:153–154
Iwasaki H, Nishiwaki T, Kitayama Y, Nakajima M, Kondo T (2002) KaiA-stimulated KaiC phosphorylation in circadian timing loops in cyanobacteria. Proc Natl Acad Sci USA 99:15788–15793
Kaye FJ, Kratzke RA, Gerster JL, Horowitz JM (1990) A single amino acid substitution results in a retinoblastoma protein defective in phosphorylation and oncoprotein binding. Proc Natl Acad Sci USA 87:6922–6926
Kodama H, Ito M, Ohnishi N, Suzuka I, Komamine A (1991) Molecular cloning of the gene for plant proliferating-cell nuclear antigen and expression of this gene during the cell cycle in synchronized cultures of Catharanthus roseus cells. Eur J Biochem 197:495–503
Kong L–J, Orozco BM, Roe JL, Nagar S, Ou S, Feiler HS, Durfee T, Miller AB, Gruissem W, Robertson D, Hanley-Bowdoin L (2000) A geminivirus replication protein interacts with the retinoblastoma protein through a novel domain to determine symptoms and tissue specificity of infection in plants. EMBO J 19:3485–3495
Lin JY, DeCaprio JA (2003) SV40 large T antigen promotes dephosphorylation of p130. J Biol Chem 278:46482–46487
Miao G-H, Hong Z, Verma DPS (1993) Two functional soybean genes encoding p34cdc2 protein kinases are regulated by different plant developmental pathways. Proc Natl Acad Sci USA 90:943–947
Nakagami H, Sekine M, Murakami H, Shinmyo A (1999) Tobacco retinoblastoma-related protein phosphorylated by a distinct cyclin-dependent kinase complex with Cdc2/cyclin D in vitro. Plant J 18:243–252
Nakagami H, Kawamura K, Sugisaka K, Sekine M, Shinmyo A (2002) Phosphorylation of retinoblastoma-related protein by the cyclinD/cyclin-dependent kinase complex is activated at the G1/S-phase transition in tobacco. Plant Cell 14:1847–1857
Ramirez-Parra E, Xie Q, Boniotti MB, Gutierrez C (1999) The cloning of plant E2F, a retinoblastoma-binding protein, reveals unique and conserved features with animal G1/S regulators. Nucleic Acids Res 27:3527–3533
Sabelli PA, Dante RA, Leiva-Neto JT, Jung R, Gordon-Kamm WJ, Larkins BA (2005) RBR3, a member of the retinoblastoma-related family from maize, is regulated by the RBR1/E2F pathway. Proc Natl Acad Sci USA 102:13005–13012
Sauter M (1997) Differential expression of a CAK (cdc2-activating kinase)-like protein kinase, cyclins and cdc2 genes from rice during the cell cycle and in response to gibberellin. Plant J 11:181–190
Sekine M, Ito M, Uemukai K, Maeda Y, Nakagami H, Shinmyo A (1999) Isolation and characterization of the E2F-like gene in plants. FEBS Lett 460:117–122
Sherr CJ (2004) Principles of tumor suppression. Cell 116:235–246
Shimizu S, Mori H (1998) Analysis of cycles of dormancy and growth in pea axillary buds based on mRNA accumulation patterns of cell cycle-related genes. Plant Cell Physiol 39:255–262
Shimizu-Sato S, Mori H (2001) Control of outgrowth and dormancy in axillary buds. Plant Physiol 127:1405–1413
Soni R, Carmichael JP, Shah ZH, Murray JAH (1995) A family of cyclin D homologs from plants differentially controlled by growth regulators and containing the conserved retinoblastoma protein interaction motif. Plant Cell 7:85–103
Tanaka M, Takei K, Kojima M, Sakakibara H, Mori H (2006) Auxin controls local cytokinin biosynthesis in the nodal stem in apical dominance. Plant J 45:1028–1036
Tedesco D, Lukas J, Reed SI (2002) The pRb-related protein p130 is regulated by phosphorylation-dependent proteolysis via the protein-ubiquitin ligase SCFSkp2. Genes Dev 16:2946–2957
Thimann KV, Skoog F (1934) On the inhibition of bud development and other functions of growth substance in Vicia faba. Proc R Soc London Ser B 114:317–339
Wildwater M, Campilho A, Perez-Perez JM, Heidstra R, Blilou I, Korthout H, Chatterjee J, Mariconti L, Gruissem W, Scheres B (2005) The RETINOBLASTOMA-RELATED gene regulates stem cell maintenance in Arabidopsis roots. Cell 123:1337–1349
Xie Q, Sanz-Burgos AP, Hannon GJ, Gutierrez C (1996) Plant cells contain a novel member of the retinoblastoma family of growth regulatory proteins. EMBO J 15:4900–4908
Acknowledgements
This work was supported in part by Grants-in Aid to H.M. from the Japan Ministry of Education, Sciences, Sports, and Culture (No. 17027013, 17051014, and 19039015). S.S.-S. has received Research Fellowships of the Japan Society for the Promotion of Science for Young Scientists.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License (https://creativecommons.org/licenses/by-nc/2.0), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Shimizu-Sato, S., Ike, Y. & Mori, H. PsRBR1 encodes a pea retinoblastoma-related protein that is phosphorylated in axillary buds during dormancy-to-growth transition. Plant Mol Biol 66, 125–135 (2008). https://doi.org/10.1007/s11103-007-9257-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11103-007-9257-5