Abstract
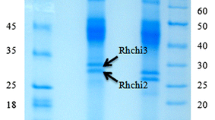
We previously isolated a Brassica juncea cDNA encoding BjCHI1, a novel chitinase with two chitin-binding domains. Synthesis of its mRNA is induced by wounding, methyl jasmonate treatment, Aspergillus niger infection and caterpillar Pieris rapae feeding, suggesting that the protein has a role in defense. In that it possesses two chitin-binding domains, BjCHI1 resembles the precursor of Urtica dioica agglutinin but unlike that protein, BjCHI1 retains its chitinase catalytic domain after post-translational processing. To explore the properties of multi-domain BjCHI1, we have expressed recombinant BjCHI1 and two derivatives, which lack one (BjCHI2) or both (BjCHI3) chitin-binding domains, as secreted proteins in Pichia pastoris. Recombinant BjCHI1 and BjCHI2, showed apparent molecular masses on SDS-PAGE larger than calculated, and could be deglycosylated using α-mannosidase. Recombinant BjCHI3, without the proline/threonine-rich linker region containing predicted O-glycosylation sites, did not appear to be processed by α-mannosidase. BjCHI1’s ability to agglutinate rabbit erythrocytes is unique among known chitinases. Both chitin-binding domains are essential for agglutination; this property is absent in recombinant BjCHI2 and BjCHI3. To identify potential catalytic residues, we generated site-directed mutations in recombinant BjCHI3. Mutation E212A showed the largest effect, exhibiting 0 of wild-type specific activity. H211N and R361A resulted in considerable (>91) activity loss, implying these charged residues are also important in catalysis. E234A showed 36 retention of activity and substitution Y269D, 50. The least affected mutants were E349A and D360A, with 73 and 68 retention, respectively. Like Y269, E349 and D360 are possibly involved in substrate binding rather than catalysis.
Similar content being viewed by others
References
Andersen, M. D., Jensen, A., Robertus, J. D., Leah, R. and Skriver, K. 1997. Heterologous expression and character-ization of wild-type and mutant forms of a 26 kDa endochitinase from barley (Hordeum vulgare L. ). Biochem. J. 322: 815–822.
Arakane, Y., Zhu, Q., Matsumiya, M., Muthukrishnan, S. and Kramer, K. J. 2003. Properties of catalytic, linker and chitin-binding domains of insect chitinase. Insect Biochem. Mol. Biol. 33: 631–648.
Beintema, J. J. and Peumans, W. J. 1992. The primary structure of stinging nettle (Urtica dioica )agglutinin, a two-domain member of the hevein family. FEBS Lett. 299: 131–134.
Benson, D. A., Karsch-Mizrachi, I., Lipman, D. J., Ostell, J. and Wheeler, D. L. 2003. GenBank. Nucleic Acids Res. 31: 23–27.
Berman, H. M., Westbrook, J., Feng, Z., Gilliland, G., Bhat, T. N., Weissig, H., Shindyalov, I. N. and Bourne, P. E. 2000. The Protein Data Bank. Nucleic Acids Res. 28: 235–242.
Boller, T. 1985. Induction of hydrolases as a defense reaction against pathogens. In: J. L. Key and T. Kosuge (Eds. ), Cellular and Molecular Biology of Plant Stress. Alan R. Liss, New York, pp. 247–262.
Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72: 248–254. aiBrameld, K. A. and Goddard, W. A., III. 1998. The role of enzyme distortion in the single displacement mechanism of family 19 chitinases. Proc. Natl. Acad. Sci. USA 95: 4276–4281.
Bretthauer, R. K. 2003. Genetic engineering of Pichia pastoris to humanize N-glycosylation of proteins. Trends Plant Sci. 21: 459–462.
Broekaert, W. F., Van Parijs, J., Leyns, F., Joos, H. and Peumans, W. J. 1989. A chitin-binding lectin from stinging nettle rhizomes with antifungal properties. Science 245: 1100–1102.
Broglie, K., Chet, I., Holliday, M., Cressman, R., Biddle, P., Knowlton, S., Mauvais, C. J. and Broglie, R. 1991. Transgenic plants with enhanced resistance to the fungal pathogen Rhizoctonia solani. Science 254: 1194–1197.
Chrispeels, M. J. and Raikhel, N. V. 1991. Lectins, lectin genes, and their role in plant defense. Plant Cell 3: 1–9.
Chye, M.-L., Zhao, K.-J., He, Z.-M., Ramalingam, S. and Fung, K.-L. 2004. An agglutinating chitinase with two chitin-binding domains confers fungal protection in trans-genic potato. Planta doi: 10. 1007/s00425-004-1391-6.
Collinge, D. B., Kragh, K. M., Mikkelsen, J. D., Nielsen, K. K., Rasmussen, U. and Vad, K. 1993. Plant chitinases. Plant J. 3: 31–40.
Does, M. P., Houterman, P. M., Dekker, H. L. and Cornelissen, B. J. 1999. Processing, targeting and antifungal activity of stinging nettle agglutinin in transgenic tobacco. Plant Physiol. 120: 421–431.
Fukamizo, T. 2000. Chitinolytic enzymes: catalysis, substrate binding, and their application. Curr. Protein Pept. Sci. 1: 105–124.
Fung, K.-L., Zhao, K.-J., He, Z.-M. and Chye, M.-L. 2002. Tobacco-expressed Brassica juncea chitinase BjCHI1 shows antifungal activity in vitro. Plant Mol. Biol. 50: 283–294.
Garcia-Casado, G., Collada, C., Allona, I., Casado, R., Pacios, L. F., Aragoncillo, C. and Gomez, L. 1998. Site-directed mutagenesis of active site residues in a class I endochitinase from chestnut seeds. Glycobiology 8: 1021–1028.
Grison, R., Grezes-Besset, B., Schneider, M., Lucante, N., Olsen, L., Leguay, J.-J. and Toppan, A. 1996. Field tolerance to fungal pathogens of Brassica napus constitutively expressing a chimeric chitinase gene. Nat. Biotechnol. 14: 643–646.
Hahn, M., Hennig, M., Schlesier, B. and Ho ¨hne, W. 2000. Structure of jack bean chitinase. Acta Crystallogr. D56: 1096–1099.
Hansen, J. E., Lund, O., Rapacki, K. and Brunak, S. 1997. O-GLYCBASE version 2. 0: a revised database of O-glycosylated proteins. Nucleic Acids Res. 25: 278–282. See http://www.ebs.dtu.dk/databases/OGLYCBASE/.
Harris, M. and Jones, T. A. 2001. Molray-a web interface between O and the POV-Ray ray tracer. Acta Crystallogr. D57: 1201–1203.
Hart, P. J., Pffuger, H. D., Monzingo, A. F., Hollis, T. and Robertus, J. D. 1995. The re ned crystal structure of an endochitinase from Hordeum vulgare L. seeds at 1. 8 A ¢ª resolution. J. Mol. Biol. 248: 402–413.
Heimo, H., Palmu, K. and Suominen, I. 1997. Expression in Pichia pastoris and puri cation of Aspergillus awamori glucoamylase catalytic domain. Protein Expr. Purif. 10: 70–79.
Henrissat, B. and Bairoch, A. 1993. New families in the classification of glycosyl hydrolases based on amino acid sequence similarities. Biochem. J. 293: 781–788.
Henrissat, B., Callebaut, I., Fabrega, S., Lehn, P., Mornon, J.-P. and Davies, G. 1995. Conserved catalytic machinery and the prediction of a common fold for several families of glycosyl hydrolases. Proc. Natl. Acad. Sci. USA 92: 7090–7094.
Holm, L. and Saunder, C. 1994. Structural similarity of plant chitinase and lysozymes from animal and phage. An evolutionary connection. FEBS Lett. 340: 129–132.
Iseli, B., Armand, S., Boller, T., Neuhaus, J.-M. and Henrissat, B. 1996. Plant chitinases use two di. erent hydrolytic mechanisms. FEBS Lett. 382: 186–188.
Iseli-Gamboni, B., Boller, T. and Neuhaus, J.-M. 1998. Mutation of either of two essential glutamates convert the catalytic domain of tobacco class I chitinase into a chitin-binding lectin. Plant Sci. 134: 45–51.
Jentoft, N. 1990. Why are proteins O-glycosylated? Trends Biochem. Sci. 15: 291–294.
Jones, T. A., Zou, J.-Y., Cowan, S. W. and Kjeldgaard, M. 1991. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A47: 110–119.
Karplus, K., Sjolander, K., Barrett, C., Cline, M., Haussler, D., Hughey, R., Holm, L. and Sander, C. 1997. Predicting protein structure using hidden Markov models. Proteins S1: 134–139.
Kleywegt, G. J., Zou, J. Y., Kjeldgaard, M. and Jones, T. A. 2001. Around O. In: M. G. Rossmann and E. Arnold (Eds. ), International Tables for Crystallography: Crystallography of Biological Macromolecules, Kluwer Academic Publishers, Dordrecht, The Netherlands, pp. 353–356.
Kramer, K. J., Corpuz, L., Choi, H. K. and Muthukrishnan, S. 1993. Sequence of a cDNA and expression of the gene encoding epidermal and gut chitinases of Manducta sexta. Insect Biochem. Mol. Biol. 23: 691–701.
Kraulis, P. 1991. Molscript: a program to produce both detailed and schematic plots of protein structures. J. Appl. Crystal-logr. 24: 946–950.
Kush, A., Goyvaerts, E., Chye, M.-L. and Chua, N.-H. 1990. Laticifer speci c gene expression of Hevea brasiliensis (rubber tree). Proc. Natl. Acad. Sci. USA 87: 1787–1790.
Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227: 680–685.
Langsford, M. L., Gilkes, N. R., Singh, B., Moser, B., Miller, R. C., Jr., Warren, R. A. and Kilburn, D. G. 1987. Glycosyl-ation of bacterial cellulases prevents proteolytic cleavage between functional domains. FEBS Lett. 225: 163–167.
Lerner, D. R. and Raikhel, N. V. 1992. The gene for stinging nettle lectin (Urtica dioica agglutinin)encodes both a lectin and a chitinase. J. Biol. Chem. 267: 11085–11091.
Letourneur, O., Gervasi, G., Gaia, S., Pages, J., Watelet, B. and Jolivet, M. 2001. Characterization of Toxoplasma gondii surface antigen I (SAG1)secreted from Pichia pastoris: 297. evidence of hyper O-glycosylation. Biotechnol. Appl. Biochem. 33: 35–45.
Lin, W., Anuratha, C. S., Datta, K., Potrykus, I., Muthu-krishnan, S. and Datta, S. K. 1995. Genetic engineering of rice for resistance to sheath blight. Biotechnology 13: 686–691.
Mauch, F., Mauch-Mani, B. and Boller, T. 1988. Antifungal hydrolases in pea tissue. Plant Physiol. 88: 936–942.
Melchers, L. S., Apotheker-deGroot, M., van der Knaap, J. A., Ponstein, A. S., Sela-Buurlage, M. B., Bol, J. F., Cornelissen, B. J. C., van den Elzen, P. J. M. and Linthorst, H. J. M. 1994. A new class of tobacco chitinases homologous to bacterial exo-chitinases displays antifungal activity. Plant J. 5: 469–480.
Neuhaus, J.-M., Fritig, B., Linthorst, H. J. M., Meins, F., Mikkelsen, J. D. and Ryals, J. 1996. A revised nomenclature for chitinase genes. Plant Mol. Biol. Rep. 14: 102–104.
O’ Riordain, G., Radauer, C., Ho. mann-Sommergruber, K., Adhami, K., Peterbauer, C. K., Blanco, C., Godnic-Cvar, J., Scheiner, O., Ebner, C. and Breiteneder, H. 2002. Cloning and molecular characterization of the Hevea brasiliensis allergen Hev b 11, a class I chitinase. Clin. Exp. All. 32: 455–462.
Peumans, W. J. and Van Damme, E. J. M. 1995. Lectins as plant defense proteins. Plant Physiol. 109: 347–352.
Roberts, W. K. and Selitrenniko., C. P. 1988. Plant and bacterial chitinases differ in antifungal activity. J. Gen. Microbiol. 134: 169–176.
Sambrook, J., Fritsch, E. F. and Maniatis, T. 1989. Molecular Cloning: A Laboratory Manual, Cold Spring Harbour Laboratory Press, Cold Spring Harbour, NY.
Schlumbaum, A., Mauch, F., Vo ¨geli, U. and Boller, T. 1986. Plant chitinases are potent inhibitor of fungal growth. Nature 324: 365–367.
Scorer, C. A., Clare, J. J., McCombie, W. R., Romanos, M. A. and Sreekrishna, K. 1994. Rapid selection using G418 of high copy number transformants of Pichia pastoris for high-level foreign gene expression. Biotechnology 12: 181–184.
Soedjanaatmadja, U. M. S., Subroto, T. and Beintema, J. J. 1995. Processed products of the hevein precursor in the latex of the rubber tree (Hevea brasiliensis ). FEBS Lett. 363: 211–213.
Song, H. K. and Suh, S. W. 1996. Refined structure of the chitinase from barley seeds at 2. 0 A ¢ªresolution. Acta Crystallogr. D52: 289–298.
Stals, I., Sandra, K., Geysens, S., Contreras, R., Van Beeumen, J. and Claeyssens, M. 2004. Factors in. uencing glycosyla-tion of Trichoderma reesei cellulases. Part one: post-secretorial changes of the O-and N-glycosylation pattern of Cel7A. Glycobiology 14: 725–737.
Thompson, J. D., Higgins, D. G. and Gibson, T. J. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, positions-speci c gap penalties and weight matrix choice. Nucleic Acids Res. 22: 4673–4680.
Ueda, M., Kojima, M., Yoshikawa, T., Mitsuda, N., Araki, K., Kawaguchi, T., Miyatake, K., Arai, M. and Fukamizo, T. 2003. A novel type of family 19 chitinase from Aeromonas sp. No. 10S-24. Eur. J. Biochem. 2513–2520.
Van Damme, E. J. M., Barre, A., Rouge, P. and Peumans, W. J. 2004. Potato lectin: an updated model of a unique chimeric plant protein. Plant J. 37: 34–45.
Van Parijs, J., Broekaert, W. F., Goldstein, I. J. and Peumans W. J. 1991. Hevein: an antifungal protein from rubber-tree (Hevea brasiliensis )latex. Planta 183: 258–264.
Verburg, J. G., Rangwala, S. H., Samac, D. A., Lucknow, V. A. and Huynh, Q. K. 1993. Examination of the role of tyrosine-174 in the catalytic mechanism of the Arabidopsis thaliana-chitinase: comparison of variant chitinases gener-ated by site-directed mutagenesis and expressed in insect cells using baculovirus vectors. Arch. Biochem. Biophys. 300: 223–230.
Verburg, J. G., Smith, C. E., Lisek, C. A. and Huynh, Q. K. 1992. Identi cation of an essential tyrosine residue in the catalytic site of a chitinase isolated from Zea mays that is selectively modi ed during inactivation with 1-ethyl-3-(3-dimethlyaminopropyl)-carbodiimide. J. Biol. Chem. 267: 3886–3893.
Wirth, S. J. and Wolf, G. A. 1990. Dye-labelled substrates for the assay and detection of chitinase and lysozyme activity. J. Microbiol. Methods 12: 197–205.
Wright, H. T., Sandrasegaram, G. and Wright, C. S. 1991. Evolution of a family of N-acetylglucosamine binding proteins containing the disul de-rich domain of wheat germ agglutinin. J. Mol. Evol. 33: 283–294.
Zhao, K.-J. and Chye, M.-L. 1999. Methyl jasmonate induces expression of a novel Brassica juncea chitinase with two chitin-binding domains. Plant Mol. Biol. 40: 1009–1018.
Zhu, Q., Maher, E. A., Masoud, S., Dixon, R. A. and Lamb, C. J. 1994. Enhanced protection against fungal attack by constitutive co-expression of chitinase and glucanase genes in trangenic tobacco. Biotechnology 12: 807–812.
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Tang, C.M., Chye, ML., Ramalingam, S. et al. Functional analyses of the chitin-binding domains and the catalytic domain of Brassica juncea chitinase BjCHI1. Plant Mol Biol 56, 285–298 (2004). https://doi.org/10.1007/s11103-004-3382-1
Issue Date:
DOI: https://doi.org/10.1007/s11103-004-3382-1