Abstract
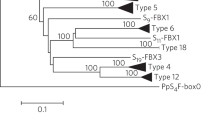
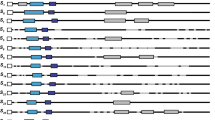
The gametophytic self-incompatibility (GSI) system in Rosaceae has been proposed to be controlled by two genes located in the S-locusan S-RNase and a recently described pollen expressed S-haplotype specific F-box gene (SFB). However, in apricot (Prunus armeniaca L.) these genes had not been identified yet. We have sequenced 21kb in total of the S-locus region in 3 different apricot S-haplotypes. These fragments contain genes homologous to the S-RNase and F-box genes found in other Prunusspecies, preserving their basic gene structure features and defined amino acid domains. The physical distance between the F-boxand the S-RNase genes was determined exactly in the S 2-haplotype (2.9kb) and inferred approximately in the S 1-haplotype (< 49kb) confirming that these genes are linked. Sequence analysis of the 5′ flanking regions indicates the presence of a conserved region upstream of the putative TATA box in the S-RNase gene. The three identified S-RNase alleles (S 1, S 2 and S 4) had a high allelic sequence diversity (75.3 amino acid identity), and the apricot F-box allelic variants (SFB1, SFB2 and SFB4) were also highly haplotype-specific (79.4 amino acid identity). Organ specific-expression was also studied, revealing that S 1- and S 2-RNases are expressed in style tissues, but not in pollen or leaves. In contrast, SFB 1 and SFB 2 are only expressed in pollen, but not in styles or leaves. Taken together, these results support these genes as candidates for the pistil and pollen S-determinants of GSI in apricot.
Similar content being viewed by others
References
Altschul, S. F., Gish, W., Miller, W., Myers, E. W. and Lipman, D. J. 1990. Basic local alignment search tool. J Mol Biol 215: 403–410.
Andrade, M. A., Gonzalez-Guzman, M., Serrano, R. and Rodrı´ guez, P. L. 2001. A combination of the F-box motif and kelch repeats defines a large Arabidopsis family of F-box proteins. Plant Mol Biol 46: 603–614.
Andrade, M. A., Ponting, C. P., Gibson, T. J. and Bork, P. 2000. Homology-based method for identification of protein repeats using statistical significance estimates. J Mol Biol 298: 521–537.
Burge, C. and Karlin, S. 1997. Prediction of complete gene structures in human genomic DNA. J Mol Biol 268: 78–94.
Burgos, L., Perez-Tornero, O., Ballester, J. and Olmos, E. 1998. Detection and inheritance of stylar ribonucleases associated with incompatibility alleles in apricot. Sex Plant Reprod 11: 153–158.
Castelo, A. T., Martins, W. and Gao, G. R. 2002. TROLLTandem Repeat Occurrence Locator. Bioinformatics 18(4): 634–636.
Channuntapipat, C., Sedgley, M. and Collins, G. G. 2001. Sequences of the cDNAs and genomic DNAs encoding the S1, S7, S8, and Sf alleles from almond, Prunus dulcis. Theor Appl Genet 103: 1115–1122.
Coleman, C. E. and Kao, T. H. 1992. The flanking regions of 2 Petunia inflata S alleles are heterogeneous and contain repetitive sequences. Plant Mol Biol 18: 725–737.
de Nettancourt, D. 1977. Incompatibility in Angiosperms. Monographs on Theoretical and Applied Genetics, 3. Springer-verlag, Berlin-Heidelberg-New York.
Doyle, J. J. and Doyle, J. L. 1987. A rapid isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull 19: 11–15.
Entani, T., Iwano, M., Shiba, H., Che, F. S., Isogai, A. and Takayama, S. 2003. Comparative analysis of the selfincompatibility (S-) locus region of Prunus mume: identifi-cation of a pollen-expressed F-box gene with allelic diversity. Genes Cells 8: 203–213.
Ficker, M., Kirch, H. H., Eijlander, R., Jacobsen, E. and Thompson, R. D. 1998. Multiple elements of the S2-RNase promoter from potato (Solanum tuberosum L. ) are required for cell type-specific expression in transgenic potato and tobacco. Mol Gen Genet 257: 132–142.
Gough, J., Karplus, K., Hughey, R. and Chothia, C. 2001. Assignment of homology to genome sequences using a library of Hidden Markov Models that represent all proteins of known structure. J Mol Biol 313(4): 903–919.
Hall, T. A. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/ NT. Nucl Acids Symp Ser 41: 95–98.
Igic, B. and Kohn, J. R. 2001. Evolutionary relationships among self-incompatibility RNases. Proc Natl Acad Sci USA 98: 13167–13171.
Ikeda, K., Igic, B., Ushijima, K., Yamane, H., Hauck, N. R., Nakano, R., Sassa, H., Iezzoni, A. F., Kohn, J. R. and Tao, R. 2004. Primary structural features of the S haplotypespecific F-box protein, SFB, in Prunus. Sex Plant Reprod 16: 235–243.
Ioerger, T. R., Gohlke, J. R., Xu, B. and Kao, T. H. 1991. Primary structural features of the self-incompatibility protein in Solanaceae. Sex Plant Reprod 4: 81–87.
Ishimizu, T., Endo, T., Yamaguchi-Kabata, Y., Nakamura, K. T., Sakiyama, F. and Norioka, S. 1998a. Identification of regions in which positive selection may operate in S-RNase of Rosaceae: Implication for S-allele-specific recognition sites in S-RNase. Febs Letters 440: 337–342.
Ishimizu, T., Shinkawa, T., Sakiyama, F. and Norioka, S. 1998b. Primary structural features of rosaceous S-RNases associated with gametophytic self-incompatibility. Plant Mol Biol 37: 931–941.
Ishizaka, T., Nakano, H., Suzuki, T. and Kitashiba, H. 2003. Characterization of the S-RNase promoters from sweet cherry (Prunus avium L. ). Genes Genet Syst 78: 191–194.
Ito, N., Phillips, S. E., Yadav, K. D. and Knowles, P. F. 1994. Crystal structure of a free radical enzyme, galactose oxidase. J Mol Biol 238: 794–814.
Kaufmann, H., Salamini, F. and Thompson, R. D. 1991. Sequence variability and gene structure at the self-incompatibility locus of Solanum tuberosum. Mol Gen Genet 226: 457–466.
Kawata, Y., Sakiyama, F., Hayashi, F. and Kyogoku, Y. 1989. Identification of two essential histidine residues of ribonuclease T2 from Aspergillus oryzae. Eur J Biochem 187: 255–262.
Kheyr-Pour, A., Bintrim, S. B., Ioerger, T. R., Remy, R., Hammond, S. A. and Kao, T. H. 1990. Sequence diversity of pistil S-proteins associated with gametophytic selfincompatibility in Nicotiana alata. Sex Plant Reprod 3: 88–97.
Lai, Z., Ma, W., Han, B., Liang, L., Zhang, Y., Hong, G. and Xue, Y. 2002. An F-box gene linked to the self-incompatibility (S) locus of Antirrhinum is expressed specifically in pollen and tapetum. Plant Mol Biol 50: 29–42.
Ma, R. C. and Oliveira, M. M. 2002. Evolutionary analysis of SRNase genes from Rosaceae species. Mol Gen Genom 267: 71–78.
Matsuura, T., Sakai, H., Unno, M., Ida, K., Sato, M., Sakiyama, F. and Norioka, S. 2001. Crystal structure at 1. 5-angstrom resolution of Pyrus pyrifolia pistil ribonuclease responsible for gametophytic self-incompatibility. J Biol Chem 276: 45261–45269.
Matton, D. P., Maes, O., Laublin, G., Xike, Q., Bertrand, C., Morse, D. and Cappadocia, M. 1997. Hypervariable domains of self-incompatibility RNases mediate allele specific pollen recognition. Plant Cell 9: 1757–1766.
Matton, D. P. S., Mau, L., Okamoto, S., Clarke, A. E. and Newbigin, E. 1995. The S-locus of Nicotiana alata: genomic organization and sequence analysis of two S-RNase alleles. Plant Mol Biol 28: 847–858.
McClure, B. A., Haring, V., Ebert, P. R., Anderson, M. A., Simpson, R. J., Sakiyama, F. and Clarke, A. E. 1989. Style self-incompatibility gene products of Nicotiana alata are ribonucleases. Nature 342: 955–957.
Nei, M. 1987. Molecular Evolutionary Genetics. Columbia Univ. Press, New York.
Nei, M. and Gojobori, T. 1986. Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol Biol Evol 3: 418–426.
Norioka, N., Katayama, H., Matsuki, T., Ishimizu, T., Takasaki, T., Nakanishi, T. and Norioka, S. 2001. Sequence comparison of the 5¢ flanking regions of Japanese pear (Pyrus pyrifolia) S-RNases associated with gametophytic self-incompatibility. Sex Plant Reprod 13: 289–291.
Rice, P., Longden, I. and Bleasby, A. 2000. EMBOSS: the European molecular biology open software suite. Trends Genet 16(6): 276–277.
Roalson, E. H. and McCubbin, A. G. 2003. S-RNases and sexual incompatibility: structure, functions, and evolutionary perspectives. Mol Phylogenet Evol 29: 490–506.
Rozas, J., Sa´ nchez-DelBarrio, J. C., Messeguer, X. and Rozas, R. 2003. DnaSP, DNA polymorphism analyses by the coalescent and other methods. Bioinformatics 19: 2496–2497.
Rozen, S. and Skaletsky, H. J. 2000. Primer3 on the WWW for general users and for biologist programmers. In: S. Krawetz, and S. Misener, (Eds. ), Bioinformatics Methods and Protocols: Methods in Molecular Biology. Humana Press, Totowa, NJ, pp. 365–386.
Salzman, R. A., Fujita, T., Zhu-Salzman, K., Hasegawa, P. M. and Bressan R. A. 1999. An improved RNA isolation method for plant tissues containing high levels of phenolic compounds or carbohydrates. Plant Mol Biol Reptr 17: 11–17.
Simpson, G. G. and Filipowitcz, W. 1996. Splicing precursors to mRNA in higher plants: mechanisms, regulation and subnuclear organization of the spliceosomal machinery. Plant Mol Biol 32: 1–41.
Sonneveld, T., Robbins, T. P., Boskovic, R., and Tobutt, K. R. 2001. Cloning of six cherry self-incompatibility alleles and development of allele-specific PCR detection. Theor Appl Genet 102: 1046–1055.
Strauss, E. C., Kobori, J. A., Siu, G. and Hood, L. E. 1986. Specific-primer-directed DNA sequencing. Anal Biochem 154(1): 353–360.
Tamura, M., Ushijima, K., Sassa, H., Hirano, H., Tao, R., Gradziel, T. M. and Dandekar, A. M. 2000. Identification of self-incompatibility genotypes of almond by allele-specific PCR analysis. Theor Appl Genet 101: 344–349.
Tao, R., Habu, T., Yamane, H. and Sugiura, A. 2002. Characterization and cDNA cloning for Sf-RNase, a molecular marker for self-compatibility, in Japanese apricot (Prunus mume). J Jpn Soc Hort Sci 71: 595–600.
Tao, R., Yamane, H. and Sugiura, A. 1999. Molecular typing of S-alleles through identification, characterization and cDNA cloning for S-RNAses in sweet cherry. J Am Soc Hort Sci 124(3): 224–233.
Thompson, J. D., Gibson, T. J., Plewniak, F., Jeanmougin, F. and Higgins, D. G. 1997. The ClustalX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25: 4876–4882.
Thompson, J. D., Higgins, D. G. and Gibson, T. J. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, positions-specific gap penalties and weight matrix choice. Nucleic Acids Res 22: 4673–4680.
Tsai, D. S., Lee, H. S., Post, L. C., Kreiling, K. M. and Kao, T. H. 1992. Sequence of an S-protein of Lycopersicon Peruvianum and comparison with other solanaceous s-proteins. Sex Plant Reprod 5: 256–263.
Ushijima K., Sassa H., Dandekar A. M., Gradziel, T. M. and Tao, R. 2003. Structural and transcriptional analysis of the self-incompatibility locus of almond: identification of a pollen-expressed F-box gene with haplotype-specific polymorphism. Plant Cell 15: 771–781.
Ushijima, K., Sassa, H., Tao, R., Yamane, H., Dandekar, A. M., Gradziel, T. M. and Hirano, H. 1998. Cloning and characterization of cDNAs encoding S-RNases from almond (Prunus dulcis): primary structural features and sequence diversity of the S-RNases in Rosaceae. Mol Gen Genet 260: 261–268.
Verica, J. A., McCubbin, A. G. and Kao, T. H. 1998. Are the hypervariable regions of S-RNases sufficient for allelespecific recognition of pollen? Plant Cell 10: 314–316.
Vilanova, S., Romero, C., Abernathy, D., Abbott, A. G., Burgos, L., Llacer, G. and Badenes, M. L. 2003. Construction and application of a bacterial artificial chromosome (BAC) library of Prunus armeniaca L. for the identification of clones linked to the self-incompatibility locus. Mol Gen Genomics 269: 685–691.
Wünsch, A. and Hormaza, J. 2004. Cloning and characterization of genomic DNA sequences of four self-incompatibility alleles in sweet cherry (Prunus avium L. ). Theor Appl Genet 108: 299–305.
Yaegaki, H., Shimada, T., Moriguchi, T., Hayama, H., Haji, T. and Yamaguchi, M. 2001. Molecular characterization of S-RNase genes and S-genotypes in the Japanese apricot (Prunus mume Sieb. et Zucc. ). Sex Plant Reprod 13: 251–257.
Yamane, H., Ikeda, K., Ushijima, K., Sassa, H. and Tao, R. 2003a. A pollen-expressed gene for a novel protein with an F-box motif that is very tightly linked to a gene for S-RNase in two species of cherry, Prunus cerasus and P. avium. Plant Cell Physiol 44: 764–769.
Yamane, H., Tao, R., Sugiura, A., Hauck, N. R. and Iezzoni, A. F. 2001. Identification and characterization of S-RNases in tetraploid sour cherry (Prunus cerasus). J Am Soc Hort Sci 126: 661–667.
Yamane, H., Ushijima, K., Sassa, H. and Tao, R. 2003b. The use of the S haplotype-specific F-box protein gene, SFB as a molecular marker for S-haplotypes and self-compatibility in Japanese apricot (Prunus mume). Theor Appl Genet 107: 1357–1361.
Yeh, R. F., Lim, L. P. and Burge, C. B. 2001. Computational inference of homologous gene structures in the human genome. Genome Res 11: 803–816.
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Romero, C., Vilanova, S., Burgos, L. et al. Analysis of the S-locus structure in Prunus armeniaca L. Identification ofS-haplotype specific S-RNase and F-box genes. Plant Mol Biol 56, 145–157 (2004). https://doi.org/10.1007/s11103-004-2651-3
Issue Date:
DOI: https://doi.org/10.1007/s11103-004-2651-3