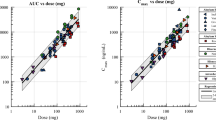
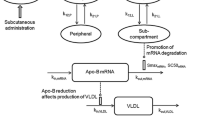
Abstract
The approval of four small interfering RNA (siRNA) products in the past few years has demonstrated unequivocally the therapeutic potential of this novel modality. Three such products (givosiran, lumasiran and inclisiran) are liver-targeted, using tris N-acetylgalactosamine (GalNAc)3 as the targeting ligand. Upon subcutaneous administration, GalNAc-conjugated siRNAs rapidly distribute into the liver via asialoglycoprotein receptor (ASGPR) mediated uptake in the hepatocytes, resulting in fast elimination from the systemic circulation. Patisiran, on the other hand, has been formulated in a lipid nanoparticle for optimal delivery to the liver. While several publications have described preclinical and clinical pharmacokinetic (PK) and pharmacodynamic (PD) results, including absorption, distribution, metabolism, and elimination (ADME) profiles in selected species as well as limited modeling efforts for siRNA therapeutics, there is no systematic review of the PK and PD models developed for these agents or work summarizing the utility and application(s) of such models in drug development and regulatory review. Here, we provide a mini-review of the current state of modeling efforts for siRNA therapeutics within the early preclinical, translational, and clinical stages of siRNA development. Diverse modeling methods including simple compartmental, mechanistic and systems PK/PD, physiologically-based PK (PBPK), population PK/PD, and dose–response-time models are introduced and reviewed. The utility of such models in development and regulatory review for siRNA therapeutics is also discussed with examples. Finally, the current knowledge gaps in mechanism of action of siRNA and resulting challenges in model development are summarized. The goal of this minireview is to trigger cross-functional discussion amongst all key stakeholders to generate key experimental datasets and align on current assumptions, model structures, and approaches to facilitate development and application of robust PK/PD models for siRNA therapeutics.

Similar content being viewed by others
References
Jorgensen R. Altered gene expression in plants due to trans interactions between homologous genes. Trends Biotechnol. 1990;8(12):340–4. https://doi.org/10.1016/0167-7799(90)90220-r.
Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391(6669):806–11. https://doi.org/10.1038/35888.
Elbashir SM, Harborth J, Lendeckel W, Yalcin A, Weber K, Tuschl T. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 2001;411(6836):494–8. https://doi.org/10.1038/35078107.
Zhang MM, Bahal R, Rasmussen TP, Manautou JE, Zhong XB. The growth of siRNA-based therapeutics: Updated clinical studies. Biochem Pharmacol. 2021;189: 114432. https://doi.org/10.1016/j.bcp.2021.114432.
Ozcan G, Ozpolat B, Coleman RL, Sood AK, Lopez-Berestein G. Preclinical and clinical development of siRNA-based therapeutics. Adv Drug Deliv Rev. 2015;87:108–19. https://doi.org/10.1016/j.addr.2015.01.007.
Gavrilov K, Saltzman WM. Therapeutic siRNA: principles, challenges, and strategies. Yale J Biol Med. 2012;85(2):187–200.
Hannon GJ. RNA interference. Nature. 2002;418(6894):244–51. https://doi.org/10.1038/418244a.
Layzer JM, McCaffrey AP, Tanner AK, Huang Z, Kay MA, Sullenger BA. In vivo activity of nuclease-resistant siRNAs. RNA. 2004;10(5):766–71. https://doi.org/10.1261/rna.5239604.
Nair JK, Willoughby JL, Chan A, Charisse K, Alam MR, Wang Q, et al. Multivalent N-acetylgalactosamine-conjugated siRNA localizes in hepatocytes and elicits robust RNAi-mediated gene silencing. J Am Chem Soc. 2014;136(49):16958–61. https://doi.org/10.1021/ja505986a.
Khvorova A, Watts JK. The chemical evolution of oligonucleotide therapies of clinical utility. Nat Biotechnol. 2017;35(3):238–48. https://doi.org/10.1038/nbt.3765.
Springer AD, Dowdy SF. GalNAc-siRNA Conjugates: Leading the Way for Delivery of RNAi Therapeutics. Nucleic Acid Ther. 2018;28(3):109–18. https://doi.org/10.1089/nat.2018.0736.
ONPATTRO [package insert]. Cambridge (MA): Alnylam Pharmacetucials, Inc; 2021.
Setten RL, Rossi JJ, Han SP. The current state and future directions of RNAi-based therapeutics. Nat Rev Drug Discov. 2019;18(6):421–46. https://doi.org/10.1038/s41573-019-0017-4.
Cullis PR, Hope MJ. Lipid Nanoparticle Systems for Enabling Gene Therapies. Mol Ther. 2017;25(7):1467–75. https://doi.org/10.1016/j.ymthe.2017.03.013.
Zhang X, Goel V, Robbie GJ. Pharmacokinetics of Patisiran, the First Approved RNA Interference Therapy in Patients With Hereditary Transthyretin-Mediated Amyloidosis. J Clin Pharmacol. 2019. https://doi.org/10.1002/jcph.1553.
Adams D, Gonzalez-Duarte A, O’Riordan WD, Yang CC, Ueda M, Kristen AV, et al. Patisiran, an RNAi Therapeutic, for Hereditary Transthyretin Amyloidosis. N Engl J Med. 2018;379(1):11–21. https://doi.org/10.1056/NEJMoa1716153.
Ayyar VS, Song D, Zheng S, Carpenter T, Heald DL. Minimal Physiologically Based Pharmacokinetic-Pharmacodynamic (mPBPK-PD) Model of N-Acetylgalactosamine-Conjugated Small Interfering RNA Disposition and Gene Silencing in Preclinical Species and Humans. J Pharmacol Exp Ther. 2021;379(2):134–46. https://doi.org/10.1124/jpet.121.000805.
GIVLAARI. [package insert]. Cambridge (MA): Alnylam Pharmaceuticals, Inc; 2021.
LEQVIO. [package insert]. East Hanover (NJ): Novartis; 2021.
OXLUMO [package insert]. Cambridge (MA): Alnylam Pharmacuticals, Inc; 2020.
US Food and Drug Administration: Population Pharmacokinectics Guidance for Industry. https://www.fda.gov/regulatory-information/search-fda-guidance-documents/population-pharmacokinetics (2022). Accessed 1 April 2022.
US Food and Drug Administration: hysiologically Based Pharmacokinetic Analyses — Format and Content Guidance for Industry. https://www.fda.gov/regulatory-information/search-fda-guidance-documents/physiologically-based-pharmacokinetic-analyses-format-and-content-guidance-industry (2018). Accessed 29 March 2022.
Fairman K, Li M, Ning B, Lumen A. Physiologically based pharmacokinetic (PBPK) modeling of RNAi therapeutics: Opportunities and challenges. Biochem Pharmacol. 2021;189: 114468. https://doi.org/10.1016/j.bcp.2021.114468.
Kanasty RL, Whitehead KA, Vegas AJ, Anderson DG. Action and reaction: the biological response to siRNA and its delivery vehicles. Mol Ther. 2012;20(3):513–24. https://doi.org/10.1038/mt.2011.294.
Bartlett DW, Davis ME. Insights into the kinetics of siRNA-mediated gene silencing from live-cell and live-animal bioluminescent imaging. Nucleic Acids Res. 2006;34(1):322–33. https://doi.org/10.1093/nar/gkj439.
Bartlett DW, Su H, Hildebrandt IJ, Weber WA, Davis ME. Impact of tumor-specific targeting on the biodistribution and efficacy of siRNA nanoparticles measured by multimodality in vivo imaging. Proc Natl Acad Sci U S A. 2007;104(39):15549–54. https://doi.org/10.1073/pnas.0707461104.
Rittenhouse KD, Johnson TR, Vicini P, Hirakawa B, Kalabat D, Yang AH, et al. RTP801 gene expression is differentially upregulated in retinopathy and is silenced by PF-04523655, a 19-Mer siRNA directed against RTP801. Invest Ophthalmol Vis Sci. 2014;55(3):1232–40. https://doi.org/10.1167/iovs.13-13449.
Zhou C, Mao Y, Sugimoto Y, Zhang Y, Kanthamneni N, Yu B, et al. SPANosomes as delivery vehicles for small interfering RNA (siRNA). Mol Pharm. 2012;9(2):201–10. https://doi.org/10.1021/mp200426h.
Mihaila R, Ruhela D, Keough E, Cherkaev E, Chang S, Galinski B, et al. Mathematical Modeling: A Tool for Optimization of Lipid Nanoparticle-Mediated Delivery of siRNA. Mol Ther Nucleic Acids. 2017;7:246–55. https://doi.org/10.1016/j.omtn.2017.04.003.
Mihaila R, Ruhela D, Galinski B, Card A, Cancilla M, Shadel T, et al. Modeling the Kinetics of Lipid-Nanoparticle- Mediated Delivery of Multiple siRNAs to Evaluate the Effect on Competition for Ago2. Mol Ther Nucleic Acids. 2019;16:367–77. https://doi.org/10.1016/j.omtn.2019.03.004.
Wei J, Jones J, Kang J, Card A, Krimm M, Hancock P, et al. RNA-induced silencing complex-bound small interfering RNA is a determinant of RNA interference-mediated gene silencing in mice. Mol Pharmacol. 2011;79(6):953–63. https://doi.org/10.1124/mol.110.070409.
Nair JK, Attarwala H, Sehgal A, Wang Q, Aluri K, Zhang X, et al. Impact of enhanced metabolic stability on pharmacokinetics and pharmacodynamics of GalNAc-siRNA conjugates. Nucleic Acids Res. 2017;45(19):10969–77. https://doi.org/10.1093/nar/gkx818.
Mager DE, Jusko WJ. Development of translational pharmacokinetic-pharmacodynamic models. Clin Pharmacol Ther. 2008;83(6):909–12. https://doi.org/10.1038/clpt.2008.52.
Shah DK, Betts AM. Towards a platform PBPK model to characterize the plasma and tissue disposition of monoclonal antibodies in preclinical species and human. J Pharmacokinet Pharmacodyn. 2012;39(1):67–86. https://doi.org/10.1007/s10928-011-9232-2.
Singh AP, Zheng X, Lin-Schmidt X, Chen W, Carpenter TJ, Zong A, et al. Development of a quantitative relationship between CAR-affinity, antigen abundance, tumor cell depletion and CAR-T cell expansion using a multiscale systems PK-PD model. MAbs. 2020;12(1):1688616. https://doi.org/10.1080/19420862.2019.1688616.
Chong S, Agarwal S, Agarwal S, Aluri KC, Arciprete M, Brown C, et al. The Nonclinical Disposition and PK/PD Properties of GalNAc-conjugated siRNA Are Highly Predictable and Build Confidence in Translation to Man. Drug Metab Dispos. 2021. https://doi.org/10.1124/dmd.121.000428.
Lee J, Goel V, Agarwal S, Melch M, Guan A, Li J, et al. Pharmacokinetic/pharmacodynamic analysis of givosiran in rats. Tenth American Conference on Pharmacometrics. Bridgewater, NJ: International Society of Pharmacometrics; 2019. p. 2688–3953.
Attarwala H, Goel V, Madigan K, Akinc A, Robbie GJ. Development of a Pharmacokinetic-Pharmacodynamic (PK-PD) Model of Fitusiran, an Investigational RNAi Therapeutic Targeting Antithrombin for the Treatment of Hemophilia in Patients With and Without Inhibitors. ISTH 2017 Congress. Berlin, Germany: Res Pract Thromb Haemost; 2017.
Agarwal S, Goel V. Clinical Pharmacology Considerations in Development of siRNA Therapeutics: Case Study of Givosiran. ASCPT 2020: Clin Pharmacol and Ther; 2020.
Sato H, Kato Y, Hayasi E, Tabata T, Suzuki M, Takahara Y, et al. A novel hepatic-targeting system for therapeutic cytokines that delivers to the hepatic asialoglycoprotein receptor, but avoids receptor-mediated endocytosis. Pharm Res. 2002;19(11):1736–44. https://doi.org/10.1023/a:1020773800358.
Ayyar VS, Jusko WJ. Transitioning from Basic toward Systems Pharmacodynamic Models: Lessons from Corticosteroids. Pharmacol Rev. 2020;72(2):414–38. https://doi.org/10.1124/pr.119.018101.
Goel V, Gosselin NH, Jomphe C, Zhang X, Marier JF, Robbie GJ. Population Pharmacokinetic-Pharmacodynamic Model of Serum Transthyretin Following Patisiran Administration. Nucleic Acid Ther. 2020;30(3):143–52. https://doi.org/10.1089/nat.2019.0841.
US Food and Drug Administration: Drug Approval Package: Onpattro (patisiran). https://www.accessdata.fda.gov/drugsatfda_docs/nda/2018/210922Orig1s000TOC.cfm (2018). Accessed 29 March 2022.
Balwani M, Sardh E, Ventura P, Peiro PA, Rees DC, Stolzel U, et al. Phase 3 Trial of RNAi Therapeutic Givosiran for Acute Intermittent Porphyria. N Engl J Med. 2020;382(24):2289–301. https://doi.org/10.1056/NEJMoa1913147.
US Food and Drug Administration: Drug Approval Package: GIVLAARI (givosiran) Injection. https://www.accessdata.fda.gov/drugsatfda_docs/nda/2019/212194Orig1s000TOC.cfm (2019). Accessed 28 March 2022.
Panhematin. [package insert]. Lebanon (NJ): Recordati Rare Diseases Inc.; 2020.
US Food and Drug Administration: Drug Approval Package: OXLUMO (lumasiran). https://www.accessdata.fda.gov/drugsatfda_docs/nda/2020/214103Orig1s000TOC.cfm (2020). Accessed 1 April 2022.
Hoppe B, Koch A, Cochat P, Garrelfs SF, Baum MA, Groothoff JW, et al. Safety, pharmacodynamics, and exposure-response modeling results from a first-in-human phase 1 study of nedosiran (PHYOX1) in primary hyperoxaluria. Kidney Int. 2022;101(3):626–34. https://doi.org/10.1016/j.kint.2021.08.015.
Bhagunde P, Ge S, Iqbal S, Mei B, Andersson S, Kanamaluru V, et al. Fitusiran Population Pharmacokinetic and Pharmacodynamic (PopPK/PD) Modeling to Support Revised Dose, Dosing Regimens & Dose Mitigation Scheme. ISTH 2021 Congress: Res Pract Thromb Haemost; 2021.
Gabrielsson J, Andersson R, Jirstrand M, Hjorth S. Dose-Response-Time Data Analysis: An Underexploited Trinity. Pharmacol Rev. 2019;71(1):89–122. https://doi.org/10.1124/pr.118.015750.
Gabrielsson J, Jusko WJ, Alari L. Modeling of dose-response-time data: four examples of estimating the turnover parameters and generating kinetic functions from response profiles. Biopharm Drug Dispos. 2000;21(2):41–52. https://doi.org/10.1002/1099-081x(200003)21:2%3c41::aid-bdd217%3e3.0.co;2-d.
Jansson-Lofmark R, Gennemark P. Inferring Half-Lives at the Effect Site of Oligonucleotide Drugs. Nucleic Acid Ther. 2018;28(6):319–25. https://doi.org/10.1089/nat.2018.0739.
Humphreys SC, Thayer MB, Campbell J, Chen WLK, Adams D, Lade JM, et al. Emerging siRNA Design Principles and Consequences for Biotransformation and Disposition in Drug Development. J Med Chem. 2020;63(12):6407–22. https://doi.org/10.1021/acs.jmedchem.9b01839.
Tomlinson B, Chow E, Chan P, Lam CWK. An evaluation of the pharmacokinetics of inclisiran in the treatment of atherosclerotic cardiovascular disease. Expert Opin Drug Metab Toxicol. 2021;17(12):1353–61. https://doi.org/10.1080/17425255.2021.2029402.
Ray KK, Wright RS, Kallend D, Koenig W, Leiter LA, Raal FJ, et al. Two Phase 3 Trials of Inclisiran in Patients with Elevated LDL Cholesterol. N Engl J Med. 2020;382(16):1507–19. https://doi.org/10.1056/NEJMoa1912387.
US Food and Drug Administration: Drug Approval Package: Leqvio (inclisiran). https://www.accessdata.fda.gov/drugsatfda_docs/nda/2022/214012Orig1s000TOC.cfm (2020). Accessed 28 March 2022.
Givlaari: EPAR - Public assessment report. https://www.ema.europa.eu/en/documents/assessment-report/givlaari-epar-public-assessment-report_en.pdf (2020). Accessed 16 June 2022.
Agarwal S, Allard R, Darcy J, Chigas S, Gu Y, Nguyen T, et al. Impact of Serum Proteins on the Uptake and RNAi Activity of GalNAc-Conjugated siRNAs. Nucleic Acid Ther. 2021;31(4):309–15. https://doi.org/10.1089/nat.2020.0919.
Basiri B, Xie F, Wu B, Humphreys SC, Lade JM, Thayer MB, et al. Introducing an In Vitro Liver Stability Assay Capable of Predicting the In Vivo Pharmacodynamic Efficacy of siRNAs for IVIVC. Mol Ther Nucleic Acids. 2020;21:725–36. https://doi.org/10.1016/j.omtn.2020.07.012.
Holford N, Ma SC, Ploeger BA. Clinical trial simulation: a review. Clin Pharmacol Ther. 2010;88(2):166–82. https://doi.org/10.1038/clpt.2010.114.
Zhang L, Gilbert PB, Capparelli E, Huang Y. Simulation-Based Pharmacokinetics Sampling Design for Evaluating Correlates of Prevention Efficacy of Passive HIV Monoclonal Antibody Prophylaxis. Statistics in Biopharmaceutical Research. 2021:1–15. https://doi.org/10.1080/19466315.2021.1919196.
Varnai P, Dave A, Farla K, Nooijen A, Petrosova L. The Evidence REVEAL Study: Exploring the Use of Real-World Evidence and Complex Clinical Trial Design by the European Pharmaceutical Industry. Clin Pharmacol Ther. 2021;110(5):1180–9. https://doi.org/10.1002/cpt.2103.
Onpattro: EPAR - Public assessment report https://www.ema.europa.eu/en/documents/assessment-report/onpattro-epar-public-assessment-report_.pdf (2018). Accessed 15 June 2022.
Oxlumo: EPAR - Public assessment report. https://www.ema.europa.eu/en/documents/assessment-report/oxlumo-epar-public-assessment-report_en.pdf (2020). Accessed 16 June 2022.
Leqvio: EPAR - Public assessment report. https://www.ema.europa.eu/en/documents/assessment-report/leqvio-epar-public-assessment-report_en.pdf (2020). Accessed 16 June 2022.
Author information
Authors and Affiliations
Contributions
All authors drafted the manuscript and contributed equally.
Corresponding author
Ethics declarations
Disclosures
This work was supported by Janssen R&D, and there is no conflict of interest to disclose.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Jeon, J.Y., Ayyar, V.S. & Mitra, A. Pharmacokinetic and Pharmacodynamic Modeling of siRNA Therapeutics – a Minireview. Pharm Res 39, 1749–1759 (2022). https://doi.org/10.1007/s11095-022-03333-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11095-022-03333-8