Abstract
Introduction
Estimation of vancomycin area under the curve (AUC) is challenging in the case of discontinuous administration. Machine learning approaches are increasingly used and can be an alternative to population pharmacokinetic (POPPK) approaches for AUC estimation.
The objectives were to train XGBoost algorithms based on simulations performed in a previous POPPK study to predict vancomycin AUC from early concentrations and a few features (i.e. patient information) and to evaluate them in a real-life external dataset in comparison to POPPK.
Patients and Methods
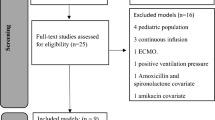
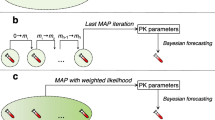
Six thousand simulations performed from 6 different POPPK models were split into training and test sets. XGBoost algorithms were trained to predict trapezoidal rule AUC a priori or based on 2, 4 or 6 samples and were evaluated by resampling in the training set and validated in the test set. Finally, the 2-sample algorithm was externally evaluated on 28 real patients and compared to a state-of-the-art POPPK model-based averaging approach.
Results
The trained algorithms showed excellent performances in the test set with relative mean prediction error (MPE)/ imprecision (RMSE) of the reference AUC = 3.3/18.9, 2.8/17.4, 1.3/13.7% for the 2, 4 and 6 samples algorithms respectively. Validation in real patient showed flexibility in sampling time post-treatment initiation and excellent performances MPE/RMSE<1.5/12% for the 2 samples algorithm in comparison to different POPPK approaches.
Conclusions
The Xgboost algorithm trained from simulation and evaluated in real patients allow accurate and precise prediction of vancomycin AUC. It can be used in combination with POPPK models to increase the confidence in AUC estimation.




Similar content being viewed by others
References
Wicha SG, Märtson A-G, Nielsen EI, Koch BCP, Friberg LE, Alffenaar J-W, et al. From therapeutic drug monitoring to model-informed precision dosing for antibiotics. Clin Pharmacol Ther. 2021;109:928–41.
Uster DW, Stocker SL, Carland JE, Brett J, Marriott DJE, Day RO, et al. A model averaging/selection approach improves the predictive performance of model-informed precision dosing: vancomycin as a case study. Clin Pharmacol Ther. 2021;109:175–83.
Chen T, Guestrin C (2016) XGBoost: A Scalable Tree Boosting System. Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining - KDD ‘16;785–94
Woillard J-B, Labriffe M, Debord J, Marquet P. Mycophenolic acid exposure prediction using machine learning. Clinical Pharmacology & Therapeutics 2021;
Woillard J-B, Labriffe M, Debord J, Marquet P. Tacrolimus exposure prediction using machine learning. Clin Pharmacol Ther 2020
Woillard J-B, Labriffe M, Prémaud A, Marquet P. Estimation of drug exposure by machine learning based on simulations from published pharmacokinetic models: the example of tacrolimus. Pharmacol Res. 2021;167:105578.
Adane ED, Herald M, Koura F. Pharmacokinetics of vancomycin in extremely obese patients with suspected or confirmed Staphylococcus aureus infections. Pharmacotherapy. 2015;35:127–39.
Revilla N, Martín-Suárez A, Pérez MP, González FM, Fernández de Gatta MDM. Vancomycin dosing assessment in intensive care unit patients based on a population pharmacokinetic/pharmacodynamic simulation. Br J Clin Pharmacol. 2010;70:201–12.
Thomson AH, Staatz CE, Tobin CM, Gall M, Lovering AM. Development and evaluation of vancomycin dosage guidelines designed to achieve new target concentrations. J Antimicrob Chemother. 2009;63:1050–7.
Roberts JA, Taccone FS, Udy AA, Vincent J-L, Jacobs F, Lipman J. Vancomycin dosing in critically ill patients: robust methods for improved continuous-infusion regimens. Antimicrob Agents Chemother. 2011;55:2704–9.
Medellín-Garibay SE, Ortiz-Martín B, Rueda-Naharro A, García B, Romano-Moreno S, Barcia E. Pharmacokinetics of vancomycin and dosing recommendations for trauma patients. J Antimicrob Chemother. 2016;71:471–9.
Mangin O, Urien S, Mainardi J-L, Fagon J-Y, Faisy C. Vancomycin pharmacokinetic and pharmacodynamic models for critically ill patients with post-sternotomy mediastinitis. Clin Pharmacokinet. 2014;53:849–61.
Wickham H, Averick M, Bryan J, Chang W, McGowan L, François R, et al. Welcome to the Tidyverse. Journal of Open Source Software. 2019;4:1686.
Kuhn M, Wickham H. Tidymodels: a collection of packages for modeling and machine learning using tidyverse principles. [Internet]. 2020. Available from: https://www.tidymodels.org
Kumar A, Roberts D, Wood KE, Light B, Parrillo JE, Sharma S, et al. Duration of hypotension before initiation of effective antimicrobial therapy is the critical determinant of survival in human septic shock. Crit Care Med. 2006;34:1589–96.
Sheiner LB, Beal S, Rosenberg B, Marathe VV. Forecasting individual pharmacokinetics. Clin Pharmacol Ther. 1979;26:294–305.
Woillard J-B, Labriffe M, Debord J, Marquet P. Tacrolimus exposure prediction using machine learning. Clin Pharmacol Ther. 2021;110:361–9.
Badillo S, Banfai B, Birzele F, Davydov II, Hutchinson L, Kam-Thong T, et al. An introduction to machine learning. Clinical Pharmacology & Therapeutics. 2020;107:871–85.
Tang B-H, Guan Z, Allegaert K, Wu Y-E, Manolis E, Leroux S, et al. Drug clearance in neonates: a combination of population pharmacokinetic modelling and machine learning approaches to improve individual prediction. Clin Pharmacokinet. 2021;60:1435–48.
Hughes JH, Keizer RJ. A hybrid machine learning/pharmacokinetic approach outperforms maximum a posteriori Bayesian estimation by selectively flattening model priors. CPT Pharmacometrics Syst Pharmacol. 2021;10:1150–60.
Acknowledgements and Disclosures
The authors thank the patients whose data were used in this study. The authors have no conflicts of interest to declare for this work.
Funding
No funding was received for this study.
Author information
Authors and Affiliations
Contributions
MB, JBW, designed the work, MB, JBW, ES, ML, PM, DWU, SGW analysis and interpret the data, MB, JBW, DWU, SGW wrote the draft.
Corresponding author
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Bououda, M., Uster, D.W., Sidorov, E. et al. A Machine Learning Approach to Predict Interdose Vancomycin Exposure. Pharm Res 39, 721–731 (2022). https://doi.org/10.1007/s11095-022-03252-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11095-022-03252-8