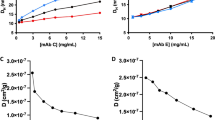
Abstract
Purpose
Reversible self-association (RSA) remains a challenge in the development of therapeutic monoclonal antibodies (mAbs). We recently analyzed the energetics of RSA for five IgG mAbs (designated as A-E) under matched conditions and using orthogonal methods. Here we examine the thermodynamics of RSA for two of the mAbs that showed the strongest evidence of RSA (mAbs C and E) to identify underlying mechanisms.
Methods
Concentration-dependent dynamic light scattering and sedimentation velocity (SV) studies were carried out for each mAb over a range of temperatures. Because self-association was weak, the SV data were globally analyzed via direct boundary fitting to identify best-fit models, accurately determine interaction energetics, and account for the confounding effects of thermodynamic and hydrodynamic nonideality.
Results
mAb C undergoes isodesmic self-association at all temperatures examined, with the energetics indicative of an enthalpically-driven reaction offset by a significant entropic penalty. By contrast, mAb E undergoes monomer-dimer self-association, with the reaction being entropically-driven and comprised of only a small enthalpic contribution.
Conclusions
Classical interpretations implicate van der Waals interactions and H-bond formation for mAb C RSA, and electrostatic interactions for mAb E. However, noting that RSA is likely coupled to additional equilibria, we also discuss the limitations of such interpretations.







Similar content being viewed by others
Abbreviations
- AUC:
-
analytical ultracentrifugation
- DLS:
-
dynamic light scattering
- mAb:
-
monoclonal antibody;
- PBS:
-
phosphate buffered saline
- RMSD:
-
root-mean-square deviation
- RSA:
-
reversible self-association
- SV:
-
sedimentation velocity
References
Ecker DM, Jones SD, Levine HL. The therapeutic monoclonal antibody market. MAbs. 2015;7(1):9–14. https://doi.org/10.4161/19420862.2015.989042.
Goswami S, Wang W, Arakawa T, Ohtake S. Developments and challenges for mAb-based therapeutics. Antibodies. 2013;2(3):452–500.
Razinkov VI, Treuheit MJ, Becker GW. Accelerated formulation development of monoclonal antibodies (mAbs) and mAb-based modalities: review of methods and tools. J Biomol Screen. 2015;20(4):468–83. https://doi.org/10.1177/1087057114565593.
Jiskoot W, Randolph TW, Volkin DB, Russell Middaugh C, Schöneich C, Winter G, et al. Protein instability and immunogenicity: roadblocks to clinical application of injectable protein delivery systems for sustained release. J Pharm Sci. 2012;101(3):946–54. https://doi.org/10.1002/jps.23018.
Shire SJ, Shahrokh Z, Liu J. Challenges in the development of high protein concentration formulations. J Pharm Sci. 2004;93(6):1390–402. https://doi.org/10.1002/jps.20079.
Philo JS, Arakawa T. Mechanisms of protein aggregation. Curr Pharm Biotechnol. 2009;10(4):348–51. https://doi.org/10.2174/138920109788488932.
Alford JR, Kendrick BS, Carpenter JF, Randolph TW. High concentration formulations of recombinant human interleukin-1 receptor antagonist: II. Aggregation kinetics. J Pharm Sci. 2008;97(8):3005–21. https://doi.org/10.1002/jps.21205.
Nishi H, Miyajima M, Nakagami H, Noda M, Uchiyama S, Fukui K. Phase separation of an IgG1 antibody solution under a low ionic strength condition. Pharm Res. 2010;27(7):1348–60. https://doi.org/10.1007/s11095-010-0125-7.
Wei JY, Bou-Assaf GM, Houde D, Weiskopf A. Technical decision-making with higher order structure data: detecting reversible concentration-dependent self-Association in a Monoclonal Antibody and a preliminary investigation to eliminate it. J Pharm Sci. 2015;104(11):3984–9. https://doi.org/10.1002/jps.24616.
Esfandiary R, Parupudi A, Casas-Finet J, Gadre D, Sathish H. Mechanism of reversible self-association of a monoclonal antibody: role of electrostatic and hydrophobic interactions. J Pharm Sci. 2015;104(2):577–86. https://doi.org/10.1002/jps.24237.
Sule SV, Cheung JK, Antochshuk V, Bhalla AS, Narasimhan C, Blaisdell S, et al. Solution pH that minimizes self-association of three monoclonal antibodies is strongly dependent on ionic strength. Mol Pharm. 2012 Apr 2;9(4):744–51. https://doi.org/10.1021/mp200448j Epub 2012 Feb 17.
Geoghegan JC, Fleming R, Damschroder M, Bishop SM, Sathish HA, Esfandiary R. Mitigation of reversible self-association and viscosity in a human IgG1 monoclonal antibody by rational, structure-guided Fv engineering. MAbs. 2016;8(5):941–50. https://doi.org/10.1080/19420862.2016.1171444.
Hopkins MM, Lambert CL, Bee JS, Parupudi A, Bain DL. Determination of interaction parameters for reversibly self-associating antibodies: a comparative analysis. J Pharm Sci. 2018;107(7):1820–30. https://doi.org/10.1016/j.xphs.2018.03.011.
Tanford C. Physical chemistry of macromolecules. New York: Wiley; 1961.
Rowe AJ. The concentration dependence of transport processes: a general description applicable to the sedimentation, translational diffusion, and viscosity coefficients of macromolecular solutes. Biopolymers. 1977;16:2595–611. https://doi.org/10.1002/bip.1977.360161202\.
Cole JL, Correia JJ, Stafford WF. The use of analytical sedimentation velocity to extract thermodynamic linkage. Biophys Chem. 2011;159(1):120–8. https://doi.org/10.1016/j.bpc.2011.05.014.
Correia JJ, Wright RT, Sherwood PJ, Stafford WF. Analysis of nonideality: insights from high concentration simulations of sedimentation velocity data. Eur Biophys J. 2020 Dec;49(8):687–700. https://doi.org/10.1007/s00249-020-01474-5. Epub 2020 Nov 6.
Esfandiary R, Hayes DB, Parupudi A, Casas-Finet J, Bai S, Samra HS, et al. A systematic multitechnique approach for detection and characterization of reversible self-association during formulation development of therapeutic antibodies. J Pharm Sci. 2013;102(9):3089–99. https://doi.org/10.1002/jps.23654.
Arora J, Hu Y, Esfandiary R, et al. Charge-mediated Fab-Fc interactions in an IgG1 antibody induce reversible self-association, cluster formation, and elevated viscosity. MAbs. 8(8):1561–74. https://doi.org/10.1080/19420862.2016.1222342.
Hu Y, Arora J, Joshi SB, Esfandiary R, Middaugh CR, Weis DD, et al. Characterization of excipient effects on reversible self-association, backbone flexibility, and solution properties of an IgG1 monoclonal antibody at high concentrations: part 1. J Pharm Sci. 2020;109(1):340–52. https://doi.org/10.1016/j.xphs.2019.06.005.
Hu Y, Toth RT 4th, Joshi SB, et al. Characterization of excipient effects on reversible self-association, backbone flexibility, and solution properties of an IgG1 monoclonal antibody at high concentrations: part 2. J Pharm Sci. 2020;109(1):353–63. https://doi.org/10.1016/j.xphs.2019.06.001.
Arora J, Hickey JM, Majumdar R, Esfandiary R, Bishop SM, Samra HS, et al. Hydrogen exchange mass spectrometry reveals protein interfaces and distant dynamic coupling effects during the reversible self-association of an IgG1 monoclonal antibody. MAbs. 2015;7(3):525–39. https://doi.org/10.1080/19420862.2015.1029217.
Hayes D, Laue T, Philo J. Program SEDNTERP: Sedimentation interpretation program. Durham, NH: University of New Hampshire;1995, www.jphilo.mailway.com/download.htm#sednterp
Stafford WF 3rd. Boundary analysis in sedimentation transport experiments: a procedure for obtaining sedimentation coefficient distributions using the time derivative of the concentration profile. Anal Biochem. 1992;203(2):295–301. https://doi.org/10.1016/0003-2697(92)90316-y.
Philo JS. Improved methods for fitting sedimentation coefficient distributions derived by time-derivative techniques. Anal Biochem. 2006;354(2):238–46. https://doi.org/10.1016/j.ab.2006.04.053.
Stafford WF, Sherwood PJ. Analysis of heterologous interacting systems by sedimentation velocity: curve fitting algorithms for estimation of sedimentation coefficients, equilibrium and kinetic constants. Biophys Chem. 2004;108(1–3):231–43. https://doi.org/10.1016/j.bpc.2003.10.028.
Claverie JM, Dreux H, Cohen R. Sedimentation of generalized systems of interacting particles. I. Solution of systems of complete Lamm equations. Biopolymers. 1975;14(8):1685–700. https://doi.org/10.1002/bip.1975.360140811.
Claverie JM. Sedimentation of generalized systems of interacting particles. III. Concentration-dependent sedimentation and extension to other transport methods. Biopolymers. 1976;15(5):843–57. https://doi.org/10.1002/bip.1976.360150504.
Sorret LL, DeWinter MA, Schwartz DK, Randolph TW. Challenges in predicting protein-protein interactions from measurements of molecular diffusivity. Biophys J. 2016 Nov 1;111(9):1831–1842. https://doi.org/10.1016/j.bpj.2016.09.018.
Correia JJ. Analysis of weight average sedimentation velocity data. Methods Enzymol. 2000;321:81–100. https://doi.org/10.1016/s0076-6879(00)21188-9.
Sontag CA, Stafford WF, Correia JJ. A comparison of weight average and direct boundary fitting of sedimentation velocity data for indefinite polymerizing systems. Biophys Chem. 2004;108(1–3):215–30. https://doi.org/10.1016/j.bpc.2003.10.029.
Correia JJ, Stafford WF. Extracting equilibrium constants from kinetically limited reacting systems. Methods Enzymol. 2009;455:419–46. https://doi.org/10.1016/S0076-6879(08)04215-8.
Creeth JM, Knight CG. On the estimation of the shape of macromolecules from sedimentation and viscosity measurements. Biochim Biophys Acta. 1965;102(2):549–58. https://doi.org/10.1016/0926-6585(65)90145-7.
Yang D, Correia JJ, Stafford WF III, et al. Weak IgG self- and hetero-association characterized by fluorescence analytical ultracentrifugation. Protein Sci. 2018;27(7):1334–48. https://doi.org/10.1002/pro.3422.
Wright RT, Hayes DB, Stafford WF, Sherwood PJ, Correia JJ. Characterization of therapeutic antibodies in the presence of human serum proteins by AU-FDS analytical ultracentrifugation. Anal Biochem. 2018 Jun;550:72–83. https://doi.org/10.1016/j.ab.2018.04.002.
Diebolder CA, Beurskens FJ, de Jong RN, Koning RI, Strumane K, Lindorfer MA, et al. Complement is activated by IgG hexamers assembled at the cell surface. Science. 2014;343(6176):1260–3. https://doi.org/10.1126/science.1248943.
de la Torre JG, Echenique Gdel R, Ortega A. Improved calculation of rotational diffusion and intrinsic viscosity of bead models for macromolecules and nanoparticles. J Phys Chem B. 2007;111(5):955–61. https://doi.org/10.1021/jp0647941.
Lin J, Lucius AL. Examination of the dynamic assembly equilibrium for E. coli ClpB. Proteins. 2015;83(11):2008–24. https://doi.org/10.1002/prot.24914.
Philo JS. Characterizing the aggregation and conformation of protein therapeutics. American Biotechnology Laboratory. 2003;21(11):22–6.
Johnson ML. Parameter correlations while curve fitting. Methods Enzymol. 2000;321:424–46. https://doi.org/10.1016/s0076-6879(00)21207-x.
Horn JR, Russell D, Lewis EA, Murphy KP. Van't Hoff and calorimetric enthalpies from isothermal titration calorimetry: are there significant discrepancies? Biochemistry. 2001;40(6):1774–8. https://doi.org/10.1021/bi002408e.
Ross PD, Subramanian S. Thermodynamics of protein association reactions: forces contributing to stability. Biochemistry. 1981;20(11):3096–102. https://doi.org/10.1021/bi00514a017.
Winzor DJ, Jackson CM. Interpretation of the temperature dependence of equilibrium and rate constants. J Mol Recognit. 2006;19(5):389–407. https://doi.org/10.1002/jmr.799.
Funding
This work was supported by MedImmune, LLC, now a member of AstraZeneca.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Disclosures of Financial Interest
The authors declare no financial interest.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
ESM 1
(DOCX 21 kb)
Rights and permissions
About this article
Cite this article
Hopkins, M.M., Parupudi, A., Bee, J.S. et al. Energetic Dissection of Mab-Specific Reversible Self-Association Reveals Unique Thermodynamic Signatures. Pharm Res 38, 243–255 (2021). https://doi.org/10.1007/s11095-021-02987-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11095-021-02987-0