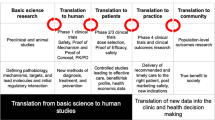
Abstract
How do we inspire new ideas that could lead to potential treatments for rare or neglected diseases, and allow for serendipity that could help to catalyze them? How many potentially good ideas are lost because they are never tested? What if those ideas could have lead to new therapeutic approaches and major healthcare advances? If a clinician or anyone for that matter, has a new idea they want to test to develop a molecule or therapeutic that they could translate to the clinic, how would they do it without a laboratory or funding? These are not idle theoretical questions but addressing them could have potentially huge economic implications for nations. If we fail to capture the diversity of ideas and test them we may also lose out on the next blockbuster treatments. Many of those involved in the process of ideation may be discouraged and simply not know where to go. We try to address these questions and describe how there are options to raising funding, how even small scale investments can foster preclinical or clinical translation, and how there are several approaches to outsourcing the experiments, whether to collaborators or commercial enterprises. While these are not new or far from complete solutions, they are first steps that can be taken by virtually anyone while we work on other solutions to build a more concrete structure for the “idea—hypothesis testing—proof of concept—translation—breakthrough pathway”.


Similar content being viewed by others
References
Sneader W. Drug discovery a history. Cheppenham: Wiley; 2005.
Nicolaou KC. The chemistry-biology-medicine continuum and the drug discovery and development process in academia. Chem Biol. 2014;21(9):1039–45.
Paul SM, Mytelka DS, Dunwiddie CT, Persinger CC, Munos BH, Lindborg SR, et al. How to improve R&D productivity: the pharmaceutical industry’s grand challenge. Nat Rev Drug Discov. 2010;9(3):203–14.
Munos BH, Orloff JJ. Disruptive innovation and transformation of the drug discovery and development enterprise. Natl Acad Med. 2016.
Baxter K, Horn E, Gal-Edd N, Zonno K, O’Leary J, Terry PF, et al. An end to the myth: there is no drug development pipeline. Sci Transl Med. 2013;5(171):171cm171.
Pariser AR, Gahl WA. Important role of translational science in rare disease innovation, discovery, and drug development. J Gen Intern Med. 2014;29 Suppl 3:S804–7.
Kerkovich D, Drew A. Designing a plan for drug discovery in rare pediatric neurodegenerative disease. Cerebrum. 2011;2011:11.
Litterman NK, Rhee M, Swinney DC, Ekins S. Collaboration for rare disease drug discovery research. F1000Res. 2014;3:261.
Wood J, Sames L, Moore A, Ekins S. Multifaceted roles of ultra-rare and rare disease patients/parents in drug discovery. Drug Discov Today. 2013;18:1043–51.
Council SE, Horvath JE. Tools for citizen-science recruitment and student engagement in your research and in your classroom. J Microbiol Biol Educ. 2016;17(1):38–40.
Wohlsen M. Biopunk: DIY scientists hack the software of life current hardcover. 2011.
Anon. scientist. Available from: http://www.scientist.com/.
Anon. Science exchange. Available from: https://www.scienceexchange.com/.
Anon. Emerald Cloud Lab. Available from: http://www.emeraldcloudlab.com/.
Ekins S, Waller CL, Bradley MP, Clark AM, Williams AJ. Four disruptive strategies for removing drug discovery bottlenecks. Drug Discov Today. 2013;18(5–6):265–71.
Siva N. Crowdfunding for medical research picks up pace. Lancet. 2014;384(9948):1085–6.
Dahlhausen K, Krebs BL, Watters JV, Ganz HH. Crowdfunding campaigns help researchers launch projects and generate outreach. J Microbiol Biol Educ. 2016;17(1):32–7.
Larkin M. How to use crowdfunding to support your research. Available from: https://www.elsevier.com/connect/how-to-use-crowdfunding-to-support-your-research.
Vachelard J, Gambarra-Soares T, Augustini G, Riul P, Maracaja-Coutinho V. A guide to scientific crowdfunding. PLoS Biol. 2016;14(2):e1002373.
Perlstein EO. Anatomy of the Crowd4Discovery crowdfunding campaign. Springerplus. 2013;2:560.
Pollastri MP. Finding new collaboration models for enabling neglected tropical disease drug discovery. PLoS Negl Trop Dis. 2014;8(7):e2866.
Pollastri MP. Improving collaborations for neglected tropical diseases. Available from: https://experiment.com/projects/improving-collaborations-for-neglected-tropical-diseases.
Riccardi G. Tuberculosis a re-emergent killer. Available from: https://universitiamo.eu/en/campaigns/tubercolosi-un-killer-riemergente.
Anon. 2016. Available from: https://www.launch.umd.edu/project/54fdb91a092065401a8df9a6.
Parish T. TB: through the looking glass. Available from: https://www.rockethub.com/projects/16030-tb-through-the-looking-glass.
Jorgensen WL. Challenges for academic drug discovery. Angew Chem Int Ed Engl. 2012;51(47):11680–4.
Anon. What are SBIR and STTR programs? Available from: http://grants.nih.gov/grants/funding/sbir.htm.
Dahlin JL, Inglese J, Walters MA. Mitigating risk in academic preclinical drug discovery. Nat Rev Drug Discov. 2015;14(4):279–94.
Sames L, Moore A, Arnold R, Ekins S. Recommendations to enable drug development for inherited neuropathies: Charcot-Marie-Tooth and Giant Axonal Neuropathy. F1000Res. 2014;3:83.
Ekins S, Wood J. Incentives for starting small companies focused on rare and neglected diseases. Pharm Res. 2016;33:809–15.
Ekins S, Hohman M, Bunin BA. Pioneering use of the cloud for development of the collaborative drug discovery (cdd) database. In: Ekins S, Hupcey MAZ, Williams AJ, editors. Collaborative computational technologies for biomedical research. Hoboken: Wiley; 2011.
LeCun Y, Bengio Y, Hinton G. Deep learning. Nature. 2015;521(7553):436–44.
Kandaswamy C, Silva LM, Alexandre LA, Santos JM. High-content analysis of breast cancer using single-cell deep transfer learning. J Biomol Screen. 2016;21(3):252–9.
Dunn SJ, Nathke IS, Osborne JM. Computational models reveal a passive mechanism for cell migration in the crypt. PLoS One. 2013;8(11):e80516.
Patel B, Gauvin R, Absar S, Gupta V, Gupta N, Nahar K, et al. Computational and bioengineered lungs as alternatives to whole animal, isolated organ, and cell-based lung models. Am J Physiol Lung Cell Mol Physiol. 2012;303(9):L733–47.
Roberts PA, Gaffney EA, Luthert PJ, Foss AJ, Byrne HM. Mathematical and computational models of the retina in health, development and disease. Prog Retin Eye Res. 2016.
Pavlides A, Hogan SJ, Bogacz R. Computational models describing possible mechanisms for generation of excessive beta oscillations in Parkinson’s disease. PLoS Comput Biol. 2015;11(12):e1004609.
Burrowes KS, De Backer J, Smallwood R, Sterk PJ, Gut I, Wirix-Speetjens R, et al. Multi-scale computational models of the airways to unravel the pathophysiological mechanisms in asthma and chronic obstructive pulmonary disease (AirPROM). Interface Focus. 2013;3(2):20120057.
Smith N, Trayanova N. Computational models of heart disease. Drug Discov Today Dis Models. 2014;14:1–2.
Sawiak SJ, Wood NI, Carpenter TA, Morton AJ. Huntington’s disease mouse models online: high-resolution MRI images with stereotaxic templates for computational neuroanatomy. PLoS One. 2012;7(12):e53361.
Moustafa AA, Gluck MA. Computational cognitive models of prefrontal-striatal-hippocampal interactions in Parkinson’s disease and schizophrenia. Neural Netw. 2011;24(6):575–91.
Habtemariam T, Tameru B, Nganwa D, Beyene G, Ayanwale L, Robnett V. Epidemiologic modeling of HIV/AIDS: use of computational models to study the population dynamics of the disease to assess effective intervention strategies for decision-making. Adv Syst Sci Appl. 2008;8(1):35–9.
ACKNOWLEDGMENTS AND DISCLOSURES
SE owns stock in Scientist (Formerly Assay Depot). SE is the CEO of Collaborations Pharmaceuticals, Inc. and Phoenix Nest, Inc.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Funding
Joseph Stahlberg Foundation grant to Dr. Aaron McMurtray.
Rights and permissions
About this article
Cite this article
Ekins, S., Diaz, N., Chung, J. et al. Enabling Anyone to Translate Clinically Relevant Ideas to Therapies. Pharm Res 34, 1–6 (2017). https://doi.org/10.1007/s11095-016-2039-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11095-016-2039-5