Abstract
A primordial environment that hosted complex pre- or proto-biochemical activity would have been subject to random fluctuations. A relevant question is then: What might be the optimum variance of such fluctuations, such that net progress could be made towards a living system? Since lipid-based membrane encapsulation was undoubtedly a key step in chemical evolution, we used a peptide-micelle system in simulated experiments where simple micelles and peptide-stabilized micelles compete for the same amphiphilic lipid substrate. As cyclic thermal driver and energy source we used a thermochemical redox oscillator, to which the micelle reactions are coupled thermally through the activation energies. The long-time series averages taken for increasing values of the fluctuation variance show two distinct minima for simple micelles, but are smoothly increasing for complex micelles. This result suggests that the fluctuation variance is an important parameter in developing and perpetuating complexity. We hypothesize that such an environment may be self-selecting for a complex, evolving chemical system to outcompete simple or parasitic molecular structures.
Similar content being viewed by others
Introduction
The milieu for the origin of life is assumed to have been physically and chemically complex over a range of time and space scales, or ‘messy’ (Szostak 2011; Powner and Sutherland 2011; Vasas et al. 2012; Cronin 2020; Bartlett and Beckett 2019), yet, in order to develop understanding as modellers, we are often constrained to study simplified versions of processes that likely were involved in the progression from chemistry to biology (Cronin 2020). In this context we describe and interrogate here a model proto-biochemical system under proxy complex conditions.
This ‘messy’ quality of the primordial environment usually refers imprecisely to the untidiness of nature. Specific studies may invoke chemical heterogeneity and reaction network complexity (Islam and Powner 2017); others focus on the dynamics of noise, for example Krakauer and Sasaki (2002) found that environmental noise makes evolution of replicators by mutation more likely. It is also known that large-scale structure and patterns in biology emerge from small-scale fluctuations (Frank 2009). But is ‘any old mess’ sufficient for life to emerge from chemistry? This is an important problem in origin of life research, because of the potential value of more precise direction in extraterrestrial habitability searches and the design of laboratory experiments.
In previous works we have stressed the potential importance of thermal messiness, and have modelled it as Gaussian fluctuations with fixed variance incident on a prebiotic thermochemical system (Ball and Brindley 2017, 2020). Our aim here is to draw some bounds on ‘messiness’ and we define this concept more precisely with respect to a particular prebiotic system by building up a picture of the behaviour of a peptide-micelle network under Gaussian fluctuation distributions of differing variance between simulations. We find that outcomes are dependent on the fluctuation variance and locate an optimum range of the variance that favours production of peptide-stabilized micelles over unstable simple micelles.
That the origin, persistence and sustenance of life must have taken place in a far from equilibrium environment now is accepted almost universally. However, although a nonequilibrium milieu in stable steady state can maintain a status quo it offers little opportunity for life’s elaboration and incorrigible inventiveness, and there is now broad recognition in the literature that such a far from equilibrium environment at the origin of life must have functioned dynamically rather than in steady state (Wang et al. 2016; Keil et al. 2008; Kitadai and Maruyama 2018).
In this context, cyclical processes are believed to be advantageous, if not essential, for complex chemistry to evolve and persist (Bywater 2009; Spitzer 2013; Varfolomeev and Lushchekina 2014), and several plausible prebiotic scenarios involving a thermally (and possibly pH) cycling environment have attracted extensive support and investigation (Wang et al. 2016). Thermo-convective circulations in the pores of oceanic hydrothermal vents as a dynamic environment for the evolution of an ‘RNA world’ have been widely modelled and experimentally investigated, and fulfil many of the requirements (Keil et al. 2008; Kitadai and Maruyama 2018); other researchers have favoured evaporation/condensation cycles in hot spring or intertidal pools (Egel 2009), a scenario which may alleviate the putative dehydration/concentration problem of polymerisation.
Evolution from the disorganised ‘soup’ (Patel et al. 2015; Saladino et al. 2018) of the very early days of the Earth’s existence to the ‘RNA world’ inevitably must have involved many other chemical species. There is now a growing consensus that RNA, peptides and protocells most likely evolved together (Bracher 2015; Petrov et al. 2015; Islam and Powner 2017; Canavelli et al. 2019; Pascal and Chen 2019; Frenkel-Pinter et al. 2020). Deamer and co-workers (Segré et al. 2001; Deamer 2017) have postulated a ‘lipid world’, where amphiphile-made surfaces and vesicles played a key role in protocell emergence, and the prebiotic importance of lipids has received considerable experimental support (Fiore et al. 2017; Colomer et al. 2020; Joshi et al. 2021). Here we are interested in eliciting the dynamics that may have favoured these early chemical associations that led to the transition to biology, particularly the evolution of membranes and development of cellularity.
Bearing in mind these various considerations, for our simulations we have developed a model in which the chemistry occurs in far from equilibrium conditions and the energy source is a periodic thermal drive provided by a comingled redox reaction, the THP (thiosulfate-hydrogen peroxide) thermochemical oscillator. We have used the THP oscillator in earlier work as provider of the necessary dynamical drive for RNA replication (Ball and Brindley 2014, 2015) and a model prebiotic metabolism (Ball and Brindley 2020). Here we use it to drive a nonspecific lipid-peptide system that is simple enough to be demonstrative and complex (but not complicated) enough to be relevant to a prebiotic scenario. To an amphiphilic lipid solution we introduce simple peptides, which star in ‘peptide world’ or ‘metabolism first’ origins hypotheses. Peptides may have amphiphilic or lipophilic properties, and their association with and stabilization of lipid membranes is thought to have been important in ancient proto-cellular systems (Bywater 2009; Egel 2009).
The model is described in detail in the next section, followed by results and discussion, and we summarise our conclusions in the final section.
Model and Assumptions
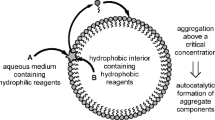
We envisage a reacting system housed in an open dissipative flow cell of volume V, which may model, for example, reactions in porous media far from equilibrium. The setup, as described and modelled below, is sketched in Fig. 1. The THP reagents hydrogen peroxide and thiosulfate, in aqueous solution, are supplied continuously to the cell where they undergo the deep oxidation,
Reaction (1) is highly exothermic, releasing some 47 kJ/g. In a non-adiabatic cell with continuous feed it may develop and sustain thermal relaxation oscillations which have been well-characterised, having been studied experimentally first by Chang and Schmitz (1975). The phenomenology is due to the high specific heat capacity of the reaction medium, which usually (although not necessarily) is aqueous solution. As reaction proceeds the cell temperature barely rises because the reaction heat excites mostly the intermolecular vibrational modes. When those modes become saturated, heat is released and the temperature spikes. Localised depletion of reactant then damps the reaction and the temperature declines, before stabilizing. Accumulation of reactant from the inflow begins the cycle again, and this behaviour may continue for as long as reactant is supplied to the cell. This, the potential energy stored during ‘charge-up’ in the quantized intermolecular modes of the medium, is the primary drive for the prebiotic reactions co-housed in the cell, described below. (Possible sources of thiosulfate and hydrogen peroxide on the early Earth were reviewed in our earlier work (Ball and Brindley 2014)).
Schematic representation of the open dissipative flow cell modelled by Eqs. (9)–(17). The cell can exchange heat with its surroundings, a thermal bath subject to fluctuations. In (a) the redox reactants of (1) are supplied to the cell, establishing thermal oscillations. In (b) the amphiphile A and small peptides B also are supplied; the thermal oscillations drive formation of active peptides P, simple micelles M and peptide-micelles \(M_P\)
The prebiotically relevant molecular system that we consider here is the formation of peptide-stabilized micellar or vesicular species, referred to henceforth as peptide-micelles. To the simulated cell (Fig. 1) we supply continuously an amphiphilic lipid or surfactant species, A, which forms micelles (or vesicles), M, autocatalytically (Bachmann et al. 1992; Ball and Haymet 2001; Morrow et al. 2019; Engwerda et al. 2015):
where (4) represents decay of M. The continuous feed supplies also an inactive small peptide B which forms, by condensation of carboxylic and ammonium groups, an active lipophilic peptide species P (Brack 1993; Sproul 2015; Lopez and Fiore 1988; Serov et al. 2020; Frenkel-Pinter et al. 2020):
The active peptide P condenses with surfactant A, or inserts itself into existing micelles, to form peptide-micelles \(M_P\), which also may decay:
In the hypothesized prebiotic setting there is no efficient enzyme catalysis and when the only operational catalysts are metal ions and surfaces, activation energies are non-negligible. Therefore, for reactions (1)–(8) the overset rate constants \(k_i (i=1\ldots 8)\) are assumed to have Arrhenius dependence on temperature T,
where the \(z_i\) are reaction frequencies, the \(E_i\) are activation energies, T is the temperature and R is the gas constant. We use mass balances as follows for evolution of the species in (1)–(8):
where x, y, a, b, p, m and \(m_p\) are dimensionless concentrations of thiosulfate, hydrogen peroxide, A, B, P, M and \(M_P\) species respectively (normalized to the input concentration of hydrogen peroxide), and \(\varphi =F/V\), F is the volumetric flow rate.
The following enthalpy balance accounts for evolution of the temperature in the cell:
where \(\bar{C}\) is the volumetric specific heat, L is the heat loss surface area multiplied by the wall thermal conductivity, subscript ‘a’ refers to ambient or environmental conditions, and \(\Delta H\) is the reaction enthalpy due to (1). (Specific reaction enthalpy contributions from (2)–(8) are assumed relatively negligible.) To simulate a thermally ‘messy’ medium, the ambient (or thermal bath) temperature \(\tilde{T}_\text {a}\) is perturbed as
where \(\bar{T}_\text {a}\) is the mean and \(\delta T_\text {a}\) is the normally distributed random fluctuation with variance \(\sigma\).
The reactant concentrations in Eqs. (10)–(16) are coupled dynamically to the temperature as it is governed by Eq. (17), for non-zero activation energies as indicated in Eq. (9). For computational and comparative purposes it is convenient to define dimensionless activation energies relative to that for the THP reaction R1, as \(\mu _i=E_i/E_1\), with numerical values given in Table 1. (In the absence of of reported experimental rate data, absolute values of the activation energies for reactions (2)–(8) were assigned on the basis of theoretical ranges for the reaction class, and dimensionless reaction frequencies (normalized to that for reaction (1)) were set equal to 9).
This system differs from others where forced thermal cycling is imposed externally, such as the model for simplified polymerase chain reaction in Barratt et al. (2010), laboratory syntheses of glycine peptides in Lahav et al. (1978) and Serov et al. (2020), and the model chemical systems in Osipovitch et al. (2009) designed to study Parrondo’s paradox. In contrast, we set values of the independent parameters L, \(\varphi\) and \(\bar{T}_\text {a}\) so that internal oscillations would develop spontaneously and be self-sustained. An advantage of internally self-consistent thermal cycling is that the system is not dependent on capricious external forcing in nature (or selective human interference in the laboratory).
The ambient stochastic fluctuations \(\delta T_\text {a}\) in Eq. (18) affect the amplitude of the thermal oscillations in the cell, as represented in Fig. 1. This is a significant shift in focus from that in Simakov and Peŕez-Mercader (2013), where stochastic fluctuations were applied to a generic thermochemical oscillator in the steady state regime to induce oscillations, and crucial in the context of prebiotic molecular complexity and evolution. Here, as discussed in the Introduction, we are interested in the effects of stochasticity on the dynamics, i.e., in the fully developed oscillatory regime.
Results and Discussion
Equations (10)–(17), using Eqs. (9) and (18), were integrated using a stiff integrator with adaptive time step. The resulting time series data were captured after more than \({10^6}\) time steps, for which numerical experiments showed that initial conditions were ‘forgotten’. Average quantities were calculated from very long time series, \(\ge 10^8\) time steps from commencing data capture.
The variance \(\sigma\) is a measure of the temperature range of the ambient stochastic fluctuation \(\delta T_\text {a}\) in Eq. (18). The major question asked in this work is: How does the magnitude of the variance affect the relative production of unstable, simple micelles M and stabilized peptide-micelles \(M_P\)?
For heuristic purposes we present first an example time series window, computed in the oscillatory regime for a moderately high input fluctuation variance \(\sigma =1.5\) (Eq. (18)), shown in Fig. 2 rendered for the temperature (a), simple micelle concentration m (b) and peptide-micelle concentration \(m_p\) (c).
Example time series window from integration of Eqs. (9)–(18) in the oscillatory regime, with \(\sigma = 1.5\). The data are rendered in terms of (a) the temperature T, (b) normalised concentration of simple micelles m (\(\times 10^3\)), and (c) normalised concentration of peptide micelles \(m_p\) (\(\times 10\))
Reaction (1) produces the oscillating temperature time trace in (a), irregularly modified by the perturbed input (Eq. (18)). The concentrations m and \(m_p\) in (b) and (c) follow the temperature. Occasional high temperature excursions, above 350 K for example, are exhibited in (a).
The fluctuations of temperature amplitude in Fig. 2a are distributed as given in the histogram in Fig. 3. This result — that normally distributed fluctuations of the ambient temperature produce a right-weighted distribution of fluctuations of the oscillation amplitude — was obtained first in Ball and Brindley (2020), but it is important to re-present it here in the context of the question posed in the title. In general the distribution in Fig. 3 implies that over time, high temperature, high activation energy synthetic reactions must be favoured over the reverse, low activation energy degradations, thus complex prebiotic processes are enabled. For the current system we may expect production of the active peptide P (reaction (5)), having the high relative activation energy \(\mu _5=1.2\) (Table 1), to lead to significant production of peptide-micelles \(M_P\) (reactions (6) and (7)) at the expense of simple micelles M (reactions (2)–(4)).
Histogram of fluctuations of the oscillation amplitude fluctuations, \(\delta T_{osc}\), of reaction (1) in the cell, produced by a normally distributed input fluctuation according to Eq. (18). The bins contain a total of 3754 cycle maxima from a continuous time series. The preponderance of high temperature excursions allows high activation energy synthetic reactions to prevail over low activation energy hydrolysis or degradation reactions
Our main results concern the the effects of the variance \(\sigma\) on production of simple micelles and peptide-micelles, and are presented in Figs. 4 and 5. For these data we took concentration averages of simple micelles and peptide-micelles, denoted \(m_\text {(av)}\) and \(m_{p\text {(av)}}\) respectively, from computed long time series for values of \(\sigma\) from 0 to 2.
Behaviour of the full system, reactions (1)–(8), where \(m_\text {(av)}\) (a) and \(m_{p\text {(av)}}\) (b) are plotted with respect to the variance \(\sigma\). In the simulations for the data in in these plots both simple micelles M and peptide-micelles \(M_P\) (b) are produced competitively. In (b) the average temperature is also plotted, with scale at right
-
The data points plotted in Fig. 4 were obtained by setting \(k_5\), \(k_6\), \(k_7\) and \(k_8\) equal to zero in Eqs. (14)–(16). This turns off production of peptide-micelles \(M_P\) and we can study the effects of the fluctuation variance on the simple micelle system, consisting of just one reactant A and one product M. The average micelle concentration \(m_\text {(av)}\) appears constant for mild fluctuations then rises after \(\sigma \approx 0.4\), becoming flatter at higher values of \(\sigma\). However, this in itself is a counter-intuitive result. Why should the concentration of simple micelles continue to increase as the fluctuations become more violent and high temperature excursions become more frequent? Since the relative activation energy for the decay (4) is lower than those for (2) and (3) (Table 1), one might naively expect \(m_\text {(av)}\) to decrease with increasing \(\sigma\). In fact, more violent fluctuations also allow for more frequent temperature excursions to very low temperatures (see Fig. 2a); the rise in \(m_\text {(av)}\) then is due to greater availability of A before it is removed by the outflow. But the gradual attenuation of this rise for \(\sigma \gtrapprox 1.2\) suggests that simple micellization is not sustainable in a violently fluctuating medium.
-
The data used for Fig. 5 are obtained from simulations of the entire peptide-micelle system in Eqs. (10)–(16), using the relative activation energies given in Table 1, for different values of the variance \(\sigma\). Here, simple micelles and peptide-micelles both are present in the cell, thus \(m_\text {(av)}\) (a) and \(m_{p\text {(av)}}\) (b) are plotted for each value of \(\sigma\). The average temperature of the cell \(T_\text {(av)}\) at each value of \(\sigma\) was also recorded, and plotted onto (b) (where the straight red lines joining the data points are purely to guide the eye—they are not interpolated or ‘fitted’ in any sense). We note that the average temperature \(T_\text {(av)}\), (b), remains constant within half a degree so reactions are governed statistically by the temperature fluctuations. Let us consider the data in Fig. 5a, b over discrete ranges of \(\sigma\).
-
\(\sigma \lessapprox 0.4\): In (b), \(m_{p\text {(av)}}\) remains nearly constant because high-temperature fluctuations are rare and consequently the active peptide P is produced very slowly from reaction (5), although in (a) there is a corresponding decline in \(m_\text {(av)}\) due to direct ‘theft’ of the amphiphile A by (6).
-
\(0.5 \lessapprox \sigma \lessapprox 0.75\): In (b) \(m_{p\text {(av)}}\) appears to increase linearly, due to increasing availability of peptide as high-temperature fluctuations become more frequent. In (a) \(m_\text {(av)}\) also rises, steeply to a maximum, despite competing for the same substrate A. This occurs because, although availability of peptide for reaction (6) is increasing, the competition still allows substantial consumption of amphiphile by (3) and (4).
-
\(0.75 \lessapprox \sigma \lessapprox 1.5\): The inevitable happens — in (a) \(m_\text {(av)}\) declines to a deep minimum while in (b) \(m_{p\text {(av)}}\) continues to rise as increasing production of active peptide by (5) allows the more stable peptide-micelles to predominate, consuming both A and M by (6) and (7).
-
\(1.5 \lessapprox \sigma < 2\): Although the scatter in this region inevitably is large, the trends in \(m_\text {(av)}\) (a) and \(m_{p\text {(av)}}\) (b) are still clear. The rise evident in \(m_\text {(av)}\) for values of \(\sigma \gtrapprox 1.7\) again reflects that violent fluctuations, counterintuitively, can allow low activation energy reactions to proceed to significant degree.
-
What do these results imply about the nature and quality of an origin of life environment? From a coarse view of the results we can say that moderate thermal stochasticity seems beneficial for chemical evolution towards more complex systems, in this case exerting an overall effect of increasing production and persistence of peptide-micelles at the expense of simple micelles. But (of course) the devil, or the spark of life, is in the detail.
Across \(\sigma \approx 1.3\)–1.7 in Fig. 5a, \(m_\text {(av)}\) is significantly depressed over that for a non-stochastic system (i.e., for \(\sigma =0\)) and for mild stochasticity. In (b) across this region \(m_{p\text {(av)}}\) continues to rise steadily, well above its value for \(\sigma =0\) and for mild stochasticity. This moderately high range of the ambient fluctuations appears to offer the greatest potential for an evolving peptide-micelle system to out-compete a simple micelle system. We may express this important observation in a different way: Over this range, the ambient fluctuations effectively dampen reaction (3) and enhance reaction (5).
However, as always with complex dynamical systems, there are tradeoffs to be made. A stable peptide-micelle system subject to high thermal fluctuations may be sacrificed to hydrolysis of the peptide (a process not included in this model), or thermal disruption of the peptide-micelles (not observed to any significant extent due to the high activation energy used for this process, (8)), or compromised by nuisance high concentrations of less stable pure micelles or parasites.
Yet, on the other hand, the non-linear effects of ambient thermal stochasticity on competing reactions engender some redundancy that can provide a fallback position, increasing the reliability of a complex molecular system, or allowing a holding pattern to be maintained. Another optimum point for \(M_P\) production would appear to be around \(\sigma \approx 0.3\)–0.4 in Fig. 5. In this region we have a local minimum in \(m_\text {(av)}\) (a) and steady, if low, \(m_{p\text {(av)}}\). This low-fluctuation regime could well act as a safe harbour for continued production of \(M_P\) when the high-fluctuation regime fails for any reason — temporary invasion of a parasitic but non-functional species perhaps. Such ‘failsafe’ redundancy is characteristic of robust living systems, and these results suggest that it may have developed earlier as a feature of prebiotic nonequilibrium chemistry. Redundancy is only possible in a complex system, and and we note that it is present already in this minimally complex one.
We know from the right-weighted distribution in Fig. 3, and from the results of Ball and Brindley (2020), that the output thermal fluctuations may favour, statistically, synthetic reactions and complexification over degradation and simplification. The new results discussed above add an important dimension to this knowledge by informing us of the effects of the variance of the ambient — or input — fluctuations, given by Eq. 18, on the reacting system. These results tell us that even though the output fluctuation distribution may be favourable, the input fluctuation variance also must be optimal for a dynamical complex molecular system to persist and grow.
Summary and Conclusions
We have shown that
-
1.
For a competitive peptide-micelle system in a stochastic thermal bath driven by a thermochemical oscillator the outputs depend non-linearly on the fluctuation variance. At moderately high fluctuation levels production of complex peptide-micelles prevails at the expense of simple micelles. Low fluctuation levels may provide a refuge for peptide-micelles to be maintained under adverse conditions.
-
2.
For a small range of medium-level fluctuation levels simple micelles may prevail over peptide-micelles. Although the literature cited in the Introduction agrees that a stochastic environment favours complexification of prebiotic chemistry, this suggests that some fluctuations are better, or worse, than others at promoting complexity. In other words, the fluctuation variance is an important parameter.
-
3.
A qualified answer to the question of whether the messiness of nature may have been actually a necessary condition for prebiotic systems to emerge and persist is affirmative and, more specifically, is that nature may have selected environments subject to fluctuations of optimal variance during the early stages of polymer selection and evolution.
It is important to note that our results are based on a specific set of activation energies which are not unreasonable for the reactions (2)–(8). But the Arrhenius temperature dependence of reaction rates (Eq. (9)) is very sensitive, as is well-known, and becomes more so the higher the activation energy. We might expect the picture in figure reffigure5 to change, perhaps dramatically, for reactions having different relative activation energies. It does, however, seem likely that the sensitivity to fluctuations exhibited in our model system will be a feature of all relevant reacting systems, thus providing nature with a rich variety of options from which to choose. This may provide an answer to the often-pondered question: ‘Why did nature select those reactions, and those reactants, for life?’ Nature simply may have selected those of which the activation energies allow complexification to prevail over simplification in a stochastic environment and competitive milieu.
In forthcoming work we investigate the effects of input stochasticity on prebiotic reactions subject to isothermal, purely chemical or pH, oscillations driven by flow kinetic energy rather than internal-mode potential energy, in recognition of the fact that such systems must have evolved from thermal to pH drive — which has persisted universally in the living cell.
Data Availability
These are available from the corresponding author.
References
Bachmann PA, Luisi PL, Lang J (1992) Autocatalytic self-replicating micelles as models for prebiotic structures. Nature 357:57–59. https://doi.org/10.1038/357057a0
Ball R, Brindley J (2014) Hydrogen peroxide thermochemical oscillator as driver for primordial RNA replication. J R Soc Interface 11:20131052. https://doi.org/10.1098/rsif.2013.1052
Ball R, Brindley J (2015) The life story of hydrogen peroxide II: a periodic pH and thermochemical drive for the RNA world. J R Soc Interface 12:20150366. https://doi.org/10.1098/rsif.2015.0366
Ball R, Brindley J (2017) Toy trains, loaded dice, and the origin of life: Dimerization on mineral surfaces under periodic drive with Gaussian inputs. R Soc Open Science 4:17041. https://doi.org/10.1098/rsos.170141
Ball R, Brindley J (2020) Anomalous thermal fluctuation distribution sustains proto-metabolic cycles and biomolecule synthesis. Phys Chem Chem Phys 22:971. https://doi.org/10.1039/c9cp05756k
Ball R, Haymet ADJ (2001) Bistability and hysteresis in self-assembling micelle systems: phenomenology and deterministic dynamics. Phys Chem Chem Phys 3:4753–4761. https://doi.org/10.1039/b104483b
Barratt C, Lapore DM, Cherubini MJ, Schwartz PM (2010) Computational models of thermal cycling in chemical systems. Int J Chemistry 2:19–27. https://doi.org/10.5539/ijc.v2n2p19
Bartlett SJ, Beckett P (2019) Probing complexity: thermodynamics and computational mechanics approaches to origins studies. R Soc Interface Focus 9:20190058. https://doi.org/10.1098/rsfs.2019.0058
Bracher P (2015) Primordial soup that cooks itself. Nat Chem 7:273–274. https://doi.org/10.1038/nchem.2219
Brack A (1993) From amino acids to prebiotic active peptides: A chemical reconstitution. Pure Appl Chem 65:1143–1151. https://doi.org/10.1351/pac199365061143
Bywater RP (2009) Membrane-spanning peptides and the origin of life. J Theor Biol 261:407–413. https://doi.org/10.1016/j.jtbi.2009.08.001
Canavelli P, Islam S, Powner MW (2019) Peptide ligation by chemoselective aminonitrile coupling in water. Nature 571:546. https://doi.org/10.1038/s41586-019-1371-4
Chang M, Schmitz RA (1975) An experimental study of oscillatory states in a stirred reactor. Chem Eng Sci 30:21–34. https://doi.org/10.1016/0009-2509(75)85112-8)
Colomer I, Borissov A, Fletcher SP (2020) Selection from a pool of self-assembling lipid replicators. Nat Commun 11:176. https://doi.org/10.1038/s41467-019-13903-x
Cronin L (2020) A new genesis for origins research? Chem 2:601–603. https://doi.org/10.1016/j.chempr.2017.04.014
Deamer D (2017) The role of lipid membranes in life’s origin. Life 7:5. https://doi.org/10.3390/life7010005
Egel R (2009) Peptide-dominated membranes preceding the genetic takeover by RNA: latest thinking on a classic controversy. BioEssays 31:1100–1109. https://doi.org/10.1002/bies.200800226
Engwerda AHJ, Southworth J, Lebedeva MA, Scanes RJH, Kukura P, Fletcher SP (2015) Coupled metabolic cycles allow out-of-equilibrium autopoietic vesicle replication. Angew Chem Int Ed 59:15450–15455. https://doi.org/10.1002/anie.202007302
Fiore M, Madanamoothoo W, Berlioz-Barbier A, Maniti O, Girard-Egrot A, Buchet R, Strazewski P (2017) Giant vesicles from rehydrated crude mixtures containing unexpected mixtures of amphiphiles formed under plausibly prebiotic conditions. Org Biomol Chem 15:4231–4240. https://doi.org/10.1039/C7OB00708F
Frank SA (2009) The common patterns of nature. J Evol Biol 22:1563–1585. https://doi.org/10.1111/j.1420-9101.2009.01775.x
Frenkel-Pinter M, Samanta M, Ashkenasy G, Leman LJ (2020) Prebiotic peptides: Molecular hubs in the origin of life. Chem Rev 120:4707–4765. https://doi.org/10.1021/acs.chemrev.9b00664
Islam S, Powner MW (2017) Prebiotic systems chemistry: Complexity overcoming clutter. Chem 2:470–501. https://doi.org/10.1016/j.chempr.2017.03.001
Joshi MP, Sawant AA, Rajamani S (2021) Spontaneous emergence of membrane-forming protoamphiphiles from a lipid-amino acid mixture under wet-dry cycles. Chem Sci 12:2970–2978. https://doi.org/10.1039/D0SC05650B
Keil L, Hartmann M, Lanzmich S, Braun D (2008) Probing of molecular replication and accumulation in shallow heat gradients through numerical simulations. Phys Chem Chem Phys 18:20153–20159. https://doi.org/10.1039/c6cp00577b
Kitadai N, Maruyama S (2018) Origins of building blocks of life: A review. Geosci Front 9:1117–1153. https://doi.org/10.1016/j.gsf.2017.07.007
Krakauer DC, Sasaki A (2002) Noisy clues to the origin of life. Proc R Soc Lond B 269:2423–2428. https://doi.org/10.1098/rspb.2002.2127
Lahav N, White D, Chang S (1978) Peptide formation in the prebiotic era: thermal condensation of glycine in fluctuating clay environments. Science 201:67–69. https://doi.org/10.1126/science.663639
Lopez A, Fiore M (1988) Investigating prebiotic protocells for a comprehensive understanding of the origins of life: A prebiotic systems chemistry perspective. Life 9:49. https://doi.org/10.3390/life9020049
Morrow SM, Colomer I, Fletcher SP (2019) A chemically fuelled self-replicator. Nat Commun 10:1011. https://doi.org/10.1038/s41467-019-08885-9
Osipovitch DC, Barratt C, Schwartz PM (2009) Systems chemistry and Parrondo’s paradox: computational models of thermal cycling. New J Chem 33:2022–2027. https://doi.org/10.1039/b900288j
Pascal R, Chen IA (2019) From soup to peptides. Nat Chem 11:763–764. https://doi.org/10.1038/s41557-019-0318-6j
Patel BH, Percivalle C, Ritson DJ, Duffy CD, Sutherland JD (2015) Common origins of RNA, protein and lipid precursors in a cyanosulfidic protometabolism. Nat Chem 7:301–307. https://doi.org/10.1038/NCHEM.2202
Petrov AS, Gulen B, Norris AM, Kovacs NA, Bernier CR, Lanier KA, Fox GE, Harvey SC, Wartell RM, Hud NV, Williams LD (2015) History of the ribosome and the origin of translation. PNAS 112:15396–15401. https://doi.org/10.1073/pnas.1509761112
Powner MW, Sutherland JD (2011) Prebiotic chemistry: a new modus operandi. Phil Trans R Soc B 366:2870–2877. https://doi.org/10.1098/rstb.2011.0134
Saladino R, Sponer JE, Sponer J, Di Mauro E (2018) Rewarming the primordial soup: Revisitations and rediscoveries in prebiotic chemistry. Chembiochem 19:22–25. https://doi.org/10.1002/cbic.201700534
Segré D, Ben-Eli D, Deamer DW, Lancet D (2001) The lipid world. Orig Life Evol Biosph 31:119–145. https://doi.org/10.1023/A:1006746807104
Serov NY, Shtyrlin VG, Khayarov KR (2020) The kinetics and mechanisms of reactions in the flow systems glycine-sodium trimetaphosphate: imidazoles: the crucial role of imidazoles in prebiotic peptide syntheses. Amino Acids 52:811–821. https://doi.org/10.1007/s00726-020-02854-z
Simakov DSA, Peŕez-Mercader J (2013) Noise induced oscillations and coherence resonance in a generic model of the nonisothermal chemical oscillator. Sci Rep 3:2404. https://doi.org/10.1038/srep02404
Spitzer J (2013) Emergence of life from multicomponent mixtures of chemicals: The case for experiments with cycling physicochemical gradients. Astrobiology 13:404–413. https://doi.org/10.1089/ast.2012.0924
Sproul G (2015) Abiogenic syntheses of lipoamino acids and lipopeptides and their prebiotic significance. Orig Life Evol Biosph 45:427–437. https://doi.org/10.1007/s11084-015-9451-44
Szostak JW (2011) An optimal degree of physical and chemical heterogeneity for the origin of life? Phil Trans R Soc B 366:2894–2901. https://doi.org/10.1098/rstb.2011.0140
Varfolomeev SD, Lushchekina SV (2014) Prebiotic synthesis and selection of macromolecules: Thermal cycling as a condition for synthesis and combinatorial selection. Geochem Int 52:1197–1206. https://doi.org/10.1134/S0016702914130102
Vasas V, Fernando C, Santos M, Kauffman S, Szathmary E (2012) Evolution before genes. Biol Direct 7:1. https://doi.org/10.1186/1745-6150-7-1
Wang GT, Tang BH, Liu Y, Gao QY, Wang ZQ (2016) The fabrication of a supra-amphiphile for dissipative self-assembly. Chem Sci 7:1151–1155. https://doi.org/10.1039/c5sc03907
Funding
This research was partially funded by Australian Research Council Future Fellowship FT0991007 (R.B.).
Author information
Authors and Affiliations
Contributions
Both authors contributed to the study conception and design. Coding, computations and data collection and processing were carried out by Rowena Ball. Both authors drafted, revised and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics Approval
Not applicable.
Conflicts of Interest/Competing Interests/Financial Interests
The authors have none to disclose.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Ball, R., Brindley, J. Does Stochasticity Favour Complexity in a Prebiotic Peptide-Micelle System?. Orig Life Evol Biosph 51, 259–271 (2021). https://doi.org/10.1007/s11084-021-09614-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11084-021-09614-3