Abstract
The 12 different types of graphs of the 8 amino acids encoded by the presumably primeval RNY code are derived. The symmetry groups of these graphs are analyzed and coincide with the corresponding values of polar requirement for each amino acid. The symmetry groups at the codon level are partially carried over as a group or subgroup at the amino acid level. Measures of centrality of the 12 graphs indicate that all amino acids were equally relevant irrespective of its chronological order of its appearance. The elimination of any amino acid would be strongly selected against and therefore the genetic code at this stage was already frozen.
Similar content being viewed by others
Introduction
Among the organic molecules that are constituents of cells, amino acids play a prominent role as building blocks of proteins which are the quintessence of the phenotypic expression. There has been clear evidence for prebiotic formation of amino acids since the experiments of Miller (Miller 1953, 1957; Miller and Orgel 1974), which involved electrical discharges in a mixture of atmospheric gases and have been shown to produce 10 of the amino acids used in modern proteins, G – Gly, A – Ala, V – Val, D – Asp, E – Glu, P –Pro, S – Ser, L – Leu, T – Thr, I – Ile plus many other organic molecules. Seven out of these 10 amino acids are encoded by the primeval genetic code composed by RNY (R-purine, Y- pyrimidine, N-any nucleotide) codons proposed by Eigen almost 30 years ago (Eigen and Schuster 1977). This slightly degenerate RNY code comprises the following amino acids: G, A, V, D, S, T, I, N – Asn. The origin and evolution of the Standard Genetic Code (SGC) has been examined by using group theory which is a branch of mathematics to determine the symmetries of an object (Coxeter 1973). In particular, it has been shown that the putative primeval RNY code can be represented in a highly symmetrical four-dimensional hypercube (José et al. 2007). It has also been shown that by frame-shift reading mistranslations and/or by transversions in the first or third nucleotide of the RNY codons the 4-dimensional hypercube replicates, together with the appearance of new amino acids, until it generates the whole SGC (José et al. 2007, 2009).
In regard to symmetries the RNY code and the whole SGC display a primitive algebraic structure known as the Four-Klein Group, which is the only non-cyclic group. More recently, it has also been shown that depending upon the ordering of the 4 nucleotides A, U, G, and C, there are 24 ways to represent the SGC (José et al. 2012) and there are only 12 ways to represent each of the corresponding phenotypic graphs of amino acids (network of amino acids as encoded by codons taking into account the structure of the genetic code (José et al. 2014)). All graphs exhibit disjoint clusters of amino acids when their polar requirement values are used (José et al. 2014). The polar requirement is a measure of chemical properties centering on the chromatographic mobility of amino acids (Woese et al. 1966). The genetic code seems to be organized so that common substitutions cause little changes in this property (Freeland and Hurst 1998).
In this work we pose the following questions: Given that the RNY code exhibit certain symmetries, do they carry over to their corresponding phenotypic graphs of the 8 primeval amino acids? Do the polar requirement values still form clusters in the networks of amino acids?
Herein, we briefly describe the main physicochemical properties of the 8 primeval amino acids. Second, we provide some algebraic definitions for understanding the type of symmetries of a graph. Third, we present the different types of primeval graphs of amino acids together with their corresponding polar requirement values. Next, we analyze the graph topologies, the symmetry groups of each of them, and we calculate the centrality measures of the 12 graphs. Finally, we briefly discuss the present findings emphasizing its value for supporting an RNY code and its phenotypic networks of amino acids (José et al. 2014).
Physicochemical Properties of the Primeval Amino Acids
Note in Fig. 1, that all amino acids are, depending on the length of its side chain, either small (G, A, V, D, S, T, N) or even tiny (A, G, S) except I. There are 4 hydrophobic (A and T, and I and V (both aliphatic)), 2 hydroxylic (S and T), and 2 polar (N acidic and D charged). Therefore, in this small set of amino acids one can already find a wide repertoire of physicochemical properties. As we will see in the section on symmetry groups, the property of polar requirement exhibits a symmetrical pattern together with the symmetries of the graphs of these 8 amino acids.
Definitions
Graph Automorphism
A graph automorphism is a function f : V(G) → V(G) that is bijective, i.e., it has a one-to-one mapping, and preserves edge-vertex connectivity, this means that for a, b ∈ V(G) if a and b are joined by an edge then f(a) and f(b) are also joined by an edge.
Given the set N = {A, U, G, C} of the 4 RNA nucleotides, the set of all the triplets is the set NNN = N3 = {xyz|x, y, z ∈ N}, that comprises the 64 codons of the SGC. Let A be the set of all the amino acids encoded by the RNY code which is a subset of N3, here dubbed N ′. The RNY codons are the vertices of a graph K, in which the edges are given by the algebraic structure of the set N3.
Now we define in the set N ′ the following equivalence relation: for x, y ∈ N ′ xℜy ⇔ x and y encode the same amino acid in A This set is now partitioned according to the amino acid that each codon encodes, and now it is straightforward to make the quotient \( \raisebox{1ex}{$\mathrm{N}^{\prime }$}\!\left/ \!\raisebox{-1ex}{$\Re $}\right.. \) This algebraic operation converts the set N ′ in a graph G where the set of vertices V(G) will be the set of amino acids present in the RNY code and the set of edges E(G) will be defined in the following manner: two vertices will be joined by an edge if there exists two codons in the graph K, that encode those amino acids that are also joined by an edge. The graph G is constructed according to the topology of the set N3 and since it can be constructed in 24 ways which are given by the permutations of the set N, but only 12 of those permutations yield to different graphs G, then we have 12 graphs G i with i ∈ {1, 2, 3, …, 12}.
Results
Graph Topology
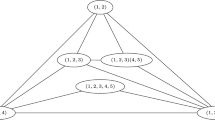
When the polar requirement scale values of each amino acid are considered, the vertices of each graph G i (see Fig. 2) are colored according to this scale and it turns out that amino acids are neatly separated into 4 major groups with 2 amino acids of the RNY code in each of these groups. Note also in Fig. 2 that the 12 graphs G i display the same core structure: two strands of four vertices each but the colors of the vertices in each strand are exactly the same. Hence we have a situation in which one amino acid of each color is present in each strand and we have the same set of amino acids on each strand regardless of the chosen graph. The strands are S 1 = {Thr, Ser, Ile, Asn}, S 2 = {Ala, Gly, Val, Asp}, where Thr and Ala are of the red group, Ser and Gly are of the green group, Ile and Val are of the blue group, and Asn and Asp are of the yellow group. Another characteristic is that for each group of polar requirement the amino acids with higher scales conform the strand S 1, and the ones with the lowest scales conform the strand S 2.
A part of the topology of a graph is given by its connectedness. If the graph is connected then it is possible to move from a fixed vertex to any other vertex following a sequence of adjacent edges. In the graphs G i , 5 (AUGC, AUCG, ACUG, ACGU, and UACG) out of the 12 are connected while the remaining 7 (AGUC, AGCU, UAGC, GAUC, GACU, and UGAC) are disconnected, and in the disconnected ones the connected components are exactly the strands S 1 and S 2.
Symmetries Groups of the 12 Graphs
The set of automorphisms of a graph does have a group structure. In Table 1, a list of the different symmetry groups of the 12 graphs of amino acids is shown: for 6 (AGUC, AGCU, UAGC, GAUC, GACU and UGAC) out of the 12 graphs G i , the group is the Klein Four-Group, for 5 (AUGC, AUCG, ACUG, ACGU, and GUAC) out of the 12, the group is the so-called Dihedral 4 group denoted by Dih4, and for only 1 (UACG) graph the group is the binary set ℤ 2 = {0, 1}. The Klein Four-Group arises on the counting of the symmetries of the rectangle which consists of two perpendicular reflections, the group Dih4 describes the complete symmetries of a square which comprises a rotation and a reflection through one diagonal of the square, and the group ℤ 2 possesses only one reflection. It is noteworthy to mention that symmetries at the codon level of the genetic code have been analyzed and the Klein Four-Group is the one that reflects these symmetries and the same group arises to describe the symmetries of graphs of the primeval amino acid since this group is also a subgroup of Dih4.
Graph Centrality Measures
The statistical centrality measures of the 12 graphs G i are shown in Table 2. The corresponding average of these estimates yielded the same value for the 2 amino acids in each polar requirement group, except for closeness in the red group composed by Ala and Thr which reflected different values. The degree is practically equal to 2 for the 8 amino acids, which means that all amino acids are uniformly connected to each other. Since all amino acids have the same value of the eigenvector this implies that all amino acids are equally relevant. Betweenness is the number of shortest paths between pairs of vertices that pass through a given vertex. In this case, betweenness measures how many times an amino acid lies on the shortest path across all the other amino acids in a graph. Closeness reflects the average distance of a vertex to all others. It can be envisaged as how long it would take to spread information from a given amino acid to all the other amino acids. Consequences of mutations and other errors in transmitting genetic information are ameliorated not only by the structure of the RNY code but also by the symmetries of the network of amino acids. Ala stands out as the amino acid with the smallest closeness centrality to the remaining amino acids, which has the largest path distances to the other amino acids.
Conclusions
The phenotypic graphs of the RNY code possess high structural symmetries which are given by the group of automorphisms present on each arrangement of the graphs G i . The symmetry group at the codon level partially carries over as a group or subgroup at the amino acid level. The Dih4 and ℤ 2 groups are new elements of symmetry in the phenotypic graphs. This is also reflected in the amino acids centrality measures which cluster each group by the scales of polar requirement regardless of the chosen measure and the type of graph. We do rarely obtain such symmetries and centrality estimates from the structure of a biological graph. However, the centrality of Ala is an outlier which will facilitate further symmetry breakings. The relevance of the 8 amino acids is the same according to its eigenvalue and given a constant degree of 2 all amino acids are evenly connected among them. According to the betweenness of the graphs any chosen amino acid can be of least length across all the remaining ones. It should not be a surprise that Ala, Gly, Val, and Asp, happen to be the most abundant amino acids formed in Miller’s experiments or found in meteorites (Miller 1953; Parker et al. 2011; Bada 2013).
The biological implications of these results are the following: At this stage of evolution of the genetic code, all amino acids were equally influential irrespective of the precise chronology of its appearance. The primeval RNY code was already frozen. All amino acids were equally relevant across all graphs except one in which the yellow group acted as a bridge between the two strands. Further evolution could only be achieved by symmetry breakings (José et al. 2007, 2009), which allowed the incorporation of new players into the graphs of amino acids. Graphs of the currently encoded 20 amino acids still show vestiges of symmetries like the ones found in this work (José et al. 2014).
References
Bada JL (2013) New insights into prebiotic chemistry from Stanley Miller’s spark discharge experiments. Chem Soc Rev 42(5):2186–2196. doi:10.1039/c3cs35433d
Coxeter HSM (1973) Regular polytopes, 3rd edn. Dover Publication Inc., New York
Eigen M, Schuster P (1977) The hypercycle. A principle of natural organization. Part A: Emergence of the hypercycle. Naturwissenschaften 64:541–565
Freeland SJ, Hurst LD (1998) The genetic code is one in a million. J Mol Evol 47:238–248
José MV, Morgado ER, Govzensky T (2007) An extended RNA code and its relationship to the standard genetic code: an algebraic and geometrical approach. Bull Math Biol 69:215–243
José MV, Goveznsky T, García JA, Bobadilla JR (2009) On the evolution of the standard genetic code: vestiges of critical scale invariance from the RNA world in current prokaryote genomes. PLoS One 4. doi:10.1371/journal.pone.0004340
José MV, Morgado ER, Govzensky T (2011) Genetic hotels for the standard genetic code: evolutionary analysis based upon novel three-dimensional algebraic models. Bull Math Biol 73:1443–1476
José MV, Morgado ER, Guimarães RC, Zamudio GS, de Farías ST, Bobadilla JR, Sosa D (2014) Three-dimensional algebraic models of the tRNA code and the 12 graphs for representing the amino acids. Life 4:341–373
Miller SL (1953) Production of amino acids under possible primitive Earth conditions. Science 117:528–529
Miller SL (1957) The mechanism of synthesis of amino acids by electric discharges. Biochim Biophys Acta 23:480–489
Miller SL, Orgel LE (1974) The origins of life on the earth. Prentice-Hall, New Jersey
Parker ET, Cleaves HJ, Dworkin JP, Glavin DP, Callahan M, Aubrey A, Lazcano A, Bada JL (2011) Primordial synthesis of amines and amino acids in a 1958 Miller H2S-rich spark discharge experiment. Proc Natl Acad Sci U S A 108(14):5526–5531. doi:10.1073/pnas.1019191108
Woese CR, Dugre DH, Saxinger WC, Dugre SA (1966) The molecular basis for the genetic code. Proc Natl Acad Sci U S A 55:966–974
Acknowledgments
MVJ was financially supported by PAPIIT-IN 224015, UNAM, México.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
José, M.V., Zamudio, G.S., Palacios-Pérez, M. et al. Symmetrical and Thermodynamic Properties of Phenotypic Graphs of Amino Acids Encoded by the Primeval RNY Code. Orig Life Evol Biosph 45, 77–83 (2015). https://doi.org/10.1007/s11084-015-9427-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11084-015-9427-4