Abstract
When Chroococcidiopsis sp. strain CCMEE 057 from the Sinai Desert and strain CCMEE 029 from the Negev Desert were exposed to space and Martian simulations in the dried status as biofilms or multilayered planktonic samples, the biofilms exhibited an enhanced rate of survival. Compared to strain CCMEE 029, biofilms of strain CCME 057 better tolerated UV polychromatic radiation (5 × 105 kJ/m2 attenuated with a 0.1 % neutral density filter) combined with space vacuum or Martian atmosphere of 780 Pa. CCMEE 029, on the other hand, failed to survive UV polychromatic doses higher than 1.5 × 103 kJ/m2. The induced damage to genomic DNA, plasma membranes and photosynthetic apparatus was quantified and visualized by means of PCR-based assays and CLSM imaging. Planktonic samples of both strains accumulated a higher amount of damage than did the biofilms after exposure to each simulation; CLSM imaging showed that photosynthetic pigment bleaching, DNA fragmentation and damaged plasma membranes occurred in the top 3–4 cell layers of both biofilms and of multilayered planktonic samples. Differences in the EPS composition were revealed by molecular probe staining as contributing to the enhanced endurance of biofilms compared to that of planktonic samples. Our results suggest that compared to strain CCMEE 029, biofilms of strain CCMEE 057 might better tolerate 1 year’s exposure in space during the next EXPOSE-R2 mission.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
In space and on the Mars surface organisms are expected to face extremely hostile environments, characterized by high vacuum (or reduced pressure), intense radiation of cosmic and solar origin and extreme temperatures (Horneck et al. 2010). The resistance of terrestrial organisms under extraterrestrial simulations and in low Earth orbit has been widely documented. After 6 years in space, multilayers of spores of Bacillus subtilis showed 70 % of survival when shielded against solar UV, while under full solar UV radiation thousands of spores survived the space journey, from an initial sample size of 108 spores, as a consequence of the protection given by the upper layers of dead spores (Horneck et al. 1994). In lichens it was suggested that the pigmented cortex provided shielding against UV radiation, thus contributing to survival after 2 weeks (Sancho et al. 2007) and 1.5 years in space (Onofri et al. 2012).
Desert strains of the cyanobacterium Chroococcidiopsis are capable of withstanding years of desiccation (Billi 2009), ionizing radiation up to 15 kGy (Billi et al. 2000) and UVC doses as high as 13 kJ/m2 (Baqué et al. 2013); possessing these key pre-requisites for survival in space and under Martian conditions, Chroococcidiopsis has been used in ground-based simulations and space exposure; a monolayer survived 10 min of simulated unattenuated Martian UV-flux (Cockell et al. 2005); a multilayer overlain by grounded sandstone survived space and Martian simulations (Billi et al. 2011), while cells augmented to an epilithic community tolerated 584 days in real space (Cockell et al. 2011).
The above described astrobiology experiments, carried out by using the European Space Agency (ESA) facilities EXPOSE and BIOPAN, as well as ground-based simulations (Rabbow et al. 2009, 2012) will be followed up by the next EXPOSE-R2 mission to be accommodated on the EXPOSE- R2 facility out of the International Space Station (ISS). This mission includes the experiments Biofilm Organisms Surfing Space (BOSS) and BIOlogy and Mars Experiment (BIOMEX) in which desert strains of the cyanobacterium Chroococcidiopsis have been included along with desiccation- tolerant organisms (de Vera et al. 2012).
In BOSS the hypothesis to be tested is: Biofilm lifestyle is better suited to support long-term survival under space and Martian conditions than is planktonic growth. In nature, most microbes occur as surface-associated communities, known as biofilms, which are encased in a self-produced matrix of extracellular polymeric substances (EPS) made of polysaccharides, proteins, nucleic acids and lipids (Flemming and Wingender 2010). Cells within biofilms differ substantially from their planktonic counterparts, particularly when facing environmental stress factors. The production of EPS is common in extreme environments, where they could assist the microbial communities to endure extremes of temperature, salinity and nutrient availability, so creating a boundary between the bacterial cell and its immediate environment (Poli et al. 2010). In addition, EPS could provide an early adaptation to minimize UV induced damage in primordial environments by facilitating cell attachment to mineral surfaces and giving protection against mineral-specific toxicity (Xu et al. 2012).
In hot and cold deserts, phototrophic biofilms occurring within porous rocks (endolithic growth) and at the interfaces between translucent stones and soil (hypolithic growth), are often dominated by cyanobacteria of the genus Chroococcidiopsis (Friedmann 1980; Billi 2012). Within these biofilms cyanobacteria are embedded in EPS as reported for endoevaporites in the Atacama Desert (Wierzchos et al. 2006; Stivaletta et al. 2012) or hypolithic communities from the Negev Desert (Grilli Caiola et al. 1996).
In the present work, resistance to space and Martian simulations of Chroococcidiopsis sp. CCMEE 029 and CCMEE 057, isolated from endolithic communities in the Negev Desert and Sinai Desert, respectively, has been investigated. Dried biofilms and dried multilayered planktonic samples were exposed to simulations planned to test organisms participating in the EXPOSE-R2 mission; these included monochromatic (254 nm, up to 10 kJ/m2) UV radiation and polychromatic (200–400 nm, 5 × 105 kJ/m2) UV radiation combined with space vacuum (2 × 10−4 Pa) or Martian atmosphere (pressure 780 Pa). The sub-cellular damage was investigated as follows: i) genome lesions by means of real-time quantitative polymerase chain reaction (qPCR) and random amplification of polymorphic DNA (RAPD); ii) plasma membrane damage by using the propidium monoazide (PMA) assay coupled to polymerase chain reaction (PCR) and by staining with the cell- impermeant molecular probe SYTOX-Green applied to confocal laser scanning microscopy (CSLM); iii): the photosynthetic pigment autofluorescence by CSLM imaging; and iv) survival by assessing the colony-forming ability. A preliminary characterization of the EPS composition of biofilms and planktonic cells was carried out by using CLSM in combination with a fluorescence- labeled lectin and lipophilic fluorescent dye.
Materials and Methods
Organisms and Sample Preparation
Chroococcidiopsis sp. CCMEE 029 (N6904) was isolated by Roseli-Ocampo Friedmann from 117 cryptoendolithic growth in sandstone, Negev Desert, Israel and CCMEE 057 (S6e) from chasmoendolithic growth in granite, Sinai Desert, Egypt. These strains are currently kept at the Department of Biology, of the University of Rome “Tor Vergata” as part of Culture Collection of Microorganisms from Extreme Environments (CCMEE) established by E. Imre Friedmann. Cyanobacteria were routinely grown at 25 °C in BG-11 medium under a photon flux density of 40 μmol/m2s1 provided by fluorescent cool-white bulbs with a 16-h/8-h light/dark cycle.
Biofilms were obtained by growing cyanobacteria on the top of BG-11 agarized medium for about 2 months; then they were left to be air-dried under routine growth conditions by removing the parafilm from the Petri dishes (85 mm); after about 15 days dried biofilms were stored in the dark at room temperature (RT). Multilayered planktonic samples were obtained by plating pellets from 2-month old liquid cultures, on the top of BG-11 agarized medium, then immediately air-dried and stored in the dark at RT. Disks were cut from dried samples to the size of the exposure carrier.
Ground-based simulations
Tests were performed at the Planetary and Space Simulation facilities (PSI) at the Institute of Aerospace Medicine (German Aerospace Center, DLR, Köln, Germany). The exposure to combined space vacuum (2 × 10−4 Pa) and 5 × 105 kJm−2 polychromatic UV radiations was carried out by placing the samples in the top and bottom positions of the carriers so that the bottom samples were shielded from radiation and exposed only to vacuum. Similarly, for the exposure to simulated Martian conditions, dried samples were collocated in carrier top and bottom positions in the presence of Mars atmosphere (argon 1.60 %, oxygen 0.15 %, nitrogen 2.70 %, carbon dioxide 95.55 % and ~370 ppm of water) of 780 Pa pressure and 5 × 105 kJ/m2 polychromatic UV radiations. A SOL2000 solar simulator was used to simulate UV exposure, which produced wavelengths greater than 200 nm. To each sample a 0.1 % neutral density (ND) filter was applied. The window material of the space simulation carrier was magnesium fluoride (λ > 110 nm), while for Martian simulation carrier suprasil was used because of its cutoff simulating present Mars UV environment (λ > 200 nm). In addition, dried samples were subjected to 10, 100, 1,000 and 10,000 J/m2 of monochromatic (254 nm) UV-radiation. All tests were performed in triplicate. Controls were kept at DLR in the dark, RT. Exposure conditions are summarized in Table 1.
Cell Integrity and DNA Damage Revealed by PCR-Based Assays
Genomic DNA was extracted from 10 mm2, in average, of Chroococcidiopsis disk samples by using the phenol-chloroform method as previously described (Billi et al. 1998), but avoiding the lysozyme and DNAseI steps, that can be used to reduce bacterial contamination in lysozyme-resistant cyanobacterial strains. DNA aliquots were normalized by readings their absorbance with an Eppendorf Biophotometer (Eppendorf, MI, Italy).
-
i)
For the PMA assay the PMA dye (Biotium, Hayward, CA) was added at a final concentration of 200 mM in DMSO to rehydrated samples. PMA enters the cells with damaged membranes, cross- links to DNA under light exposure, and thereby prevents the PCR amplification of a target gene (see below). The genomic DNA of PMA-treated cells was extracted and qPCR was performed as reported below.
-
ii)
For real-time qPCR a 1027-bp fragment of the 16S rRNA gene was used as target gene. For strain CCMEE 057 this fragment was amplified and cloned into pGEM-T vector (Promega, Italy) as reported (Stivaletta et al. 2012), while for strain CCMEE 029 it was previously obtained (Baqué et al. 2013). Known numbers of the target genes were diluted from 108 to 103 copies/μl, and standard curves were repeated for each qPCR. Amplifications were carried out in 25 μl reaction mixture consisting of 1 μl of DNA template (5 ng/μl), 12.5 of μl qPCR cocktail (iQ SYBR Green Supermix, Bio-Rad,MI,Italy)and0.05Meachof primers 16SF (5′- GGGGAATTTTCCGCAATGGGCGAAAGCCTGACGGAG-3′) and 16SR (5′- CGGGCGGTGTGTACAAGGCCCGGGAACGTATTCACC-3′). A real-time PCR detection system (iQ5, Bio-Rad, MI, Italy) was programmed to operate at 94 °C for 1 min, annealing at 60 °C for 1 min, and elongation at 72 °C for 1 min. Fluorescence measurements were recorded at the end of each annealing step. After 35 cycles a melt curve analysis was performed by recording changes in fluorescence as a function of raising the temperature from 60 °C to 95 °C in 0.5 °C per 5 s increments. All data were carried out performing n ≥ 3 qPCR protocols, each one including n = 3 replicates.
-
iii)
For RAPD analysis the primer HIP1-CA (5′-GCGATCGCCA-3′) was used in PCRs as follows: 1 cycle at 94 °C for 3 min; 30 cycles at 94 °C for 30 s, 35 °C for 30 s, 72 °C for 1 min, and 1 cycle at 72 °C for 7 min.
CLSM Imaging of Photosynthetic Pigment Autofluorescence, Live/Dead Cells and EPS Composition
Small fragments (about 2 mm2) of dried biofilms and dried multilayered planktonic samples were placed between two cover glasses with a drop of water and observed with a CLSM (Olympus Fluoview 1000 Confocal Laser Scanning System). Images were taken using a 60× objective and sequences were obtained by scanning the same x-y optical sections with a step size of 5 μm.
-
i)
The autofluorescence of the photosynthetic pigments (Chlorophyll a and phycobiliproteins) was investigated by exciting the cells with a 543-nm and a 635-nm laser and collecting the emission from 645 nm, or from 553 nm, to 800 nm emission range.
-
ii)
The integrity of the plasma membrane was assessed by using the cell-impermeant nucleic acid dye SYTOX-Green (Molecular Probes, S-7020); cells were stained with SYTOX-Green (50 μM) for 5 min in the dark. Images were taken by exciting SYTOX-Green stained cells with a 488-nm laser and collecting the emission between 510 and 530 nm.
-
iii)
To characterize the EPS composition, cells were stained for 30 min in the dark with the lectin Concanavalin A (ConA) conjugated with the fluorophore Alexa Fluor 488 (1 mg/ml, Molecular Probes C-11252) that binds α-mannopyranosyl and α-glucopyranosyl residues. Lipidic compounds were stained with the lipid-soluble green-fluorescent dye BODIPY FL C12 (100 mM, Molecular Probes D-3822). Images were taken with a 488-nm laser and by collecting the emission between 510 and 530 nm.
Results
Reduced Damage to Cell Membrane and DNA in Biofilms Compared to Planktonic Multilayers Under Space and Martian Simulations Revealed by PCR-Based Assays
For strain CCMEE 057, biofilms were more tolerant to each simulation than the multilayered planktonic samples. After exposure to space simulation (vacuum and 5 × 105 kJ/m2 polychromatic UV radiation with 0.1 % ND filter), PMA-PCR assay detected a 2-fold increase in planktonic cells with damaged membranes, and 1-fold increase in biofilms (Fig. 1). DNA damage was quantified by real-time qPCR assay as a reduction of 2.5 orders of magnitude in the copy number of the target gene in planktonic cells, and as a reduction of one order of magnitude in biofilms (Fig. 1). Under space vacuum an increase of one order of magnitude in cell membrane damage occurred in biofilms and planktonic cells, while no significant increase in DNA damage was detected (Fig. 1). Martian simulation (5 × 105 kJ/m2 polychromatic UV radiation attenuated with 0.1 % ND filter and Martian atmosphere of 780 Pa pressure) induced an increase of 2 orders and 0.5 order of magnitude in cell membrane damage in planktonic samples and biofilms, respectively (Fig. 1). Moreover, qPCR analyses of genomic DNA from biofilms and planktonic samples exposed to Martian conditions, showed a reduction of one order and 3 orders of magnitude in the amplification of the target gene, respectively (Fig. 1). Each exposed sample showed a positive colony-forming ability, except in the case of planktonic cells exposed to space conditions.
Damage to genomic DNA and plasma membranes in biofilms and multilayered planktonic samples of Chroococcidiopsis CCMEE 057 after space and Martian simulations revealed by real- time qPCR (black bars) and PMA-PCR (gray bars) Space top: vacuum (2 × 10−4 Pa ) and polychromatic UV radiation (5 × 105 kJ/m2 with 0.1 % N D 510 filter); Space bottom: vacuum (2 × 10−4 Pa); Mars top: Martian atmosphere (780 Pa) and polychromatic UV radiation (5 × 105 kJ/m2 with 0.1 % ND density filter); Mars bottom: Martian atmosphere (780 Pa)
Strain CCMEE 029, biofilms were more resistant to space vacuum and Martian atmosphere and pressure than planktonic cells: The exposure to attenuated 5 × 105 kJ/m2 UV radiation induced extensive damage to cell membranes and genomic DNA in biofilms and planktonic as revealed by PMA and qPCR assays (not shown). Planktonic and biofilm samples exposed to UV under space and Mars radiation lost their colony-forming ability.
Reduced DNA Damage in Biofilms Compared to Planktonic Multilayers Under Space and Martian Simulations Revealed by PCR-RAPD Assay
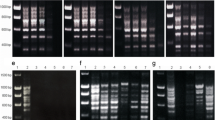
DNA damage as revealed by real-time qPCR was qualitatively confirmed by the RAPD analysis. For strain CCMEE 057 altered PCR profiles, without high-molecular weight amplicons were obtained from planktonic cells exposed to space (5 × 105 kJ/m2 polychromatic UV radiation with 0.1 % ND and vacuum) and Mars (5 × 105 kJ/m2 polychromatic UV radiation with 0.1 % ND and Martian atmosphere of 780 Pa pressure) conditions (Fig. 2a, lanes 6, 10), compared to control (Fig. 2a, lane 3). While PCR profiles virtually identical to unexposed samples were obtained from planktonic cells exposed to vacuum as well as to Martian atmosphere and pressure (Fig. 2a, lane 7, 11). No changes in PCR fingerprints were visible in biofilms after each space (Fig. 2a, lanes 4–5) or Martian simulation (Fig. 2a, lanes 8–9).
Genome damage in biofilms and multilayered planktonic samples of Chroococcidiopsis of CCMEE 057 (a) and CCMEE 029 (b) exposed to space and Martian simulations revealed by RAPD. Lanes: 2, biofilm control, 3, planktonic control, 4 biofilm space top; 5 biofilm space bottom; 6 planktonic space top, 7 planktonic space bottom, 8 biofilm Mars top; 9 biofilm Mars bottom; 10 planktonic Mars top; 11 planktonic Mars bottom. Top and bottom conditions as in Fig. 1. Lane: 1 DNA ladder
For strain CCMEE 029 planktonic cells exposed to space (5 × 105 kJ/m2 polychromatic UV radiation with 0.1 % ND filter and vacuum) and Mars (5 × 105 kJ/m2 polychromatic UV radiation with 0.1 % ND filter, Mars atmosphere and pressure) conditions exhibited PCR profiles lacking high-molecular weight amplicons (Fig. 2b, lanes 6, 10), compared to control (Fig. 2b, lanes 2–3). Under the same conditions biofilms yielded PCR-fingerprinting with a reduced abundance in PCR amplicons (Fig. 2b, lane 4, 8). Vacuum yielded a reduction in the band intensity of the PCR profiles of planktonic cells exposed to space vacuum (Fig. 2b lane 7) and no visible changes in biofilms (Fig. 2b, lane 5). The exposure to atmosphere and pressure alone did not alter the PCR fingerprints of planktonic cells (Fig. 2b, lane 11) and biofilms (Fig. 2b, lane 9).
Bleached Top Layers in Biofilms and Planktonic Multilayers Exposed to Space Simulation
CLSM imaging of biofilms of strain CCMEE 057 exposed to space simulation revealed that cells with bleached photosynthetic pigments occurred in the top 3–4 layers, whereas cells beneath had unbleached photosynthetic pigments (Fig. 3a). In dried multilayered planktonic samples, strongly bleached cells occurred in the upper layers (Fig. 3b). By contrast, unexposed, dried biofilms (Fig. 3c) and planktonic samples (not shown) revealed a red autofluorescence due to phycobiliproteins and chlorophyll a in all cell layers. Bleached top layers occurred in biofilms and planktonic samples of strain CCMEE 029 after exposure to space conditions (not shown).
Photosynthetic pigments bleaching in biofilms and multilayered planktonic samples of Chroococcidiopsis CCMEE 057 exposed to space simulation (space top). Shown is a bi-channel (543-nm and 635-nm laser) maximum-intensity projections of x-y optical sections (z step=0.53 micron) and orthogonal views in the z direction. A biofilm (a 48 μm total thickness) and multilayered planktonic sample (b, 38 μm total thickness) showing yellowish, bleached cells in the top layers and unbleached cells in the bottom layers; unbleached biofilm control (c 46 μm total thickness). t: upper layer of the sample, b: bottom layer of the sample. Scale bar = 5 μm
UVC-Induced Damage in Biofilms
The quantification of DNA damage via qPCR assay in biofilms of strain CCMEE 057 exposed to UVC doses revealed a significant decrease in the copy number of the target gene after exposure to 10 J/m2, while at higher doses (up to 10 kJ/m2) no further increase in the DNA damage was detected (Fig. 4a). When RAPD assay was performed on UVC-exposed samples no evident alterations in the PCR fingerprints occurred as compared to control (not shown).
Damage to genomic DNA and plasma membranes in biofilms of Chroococcidiopsis CCMEE 057 exposed to UVC radiation. Real-time qPCR of a target gene after 10 J/m2, 100 J/m2, 1 kJ/m2 and 10 kJ/m2 (a). CLSM image after exposure to 10 kJ/m2 showing top layer cells with damaged plasma membranes and SYTOX-green stained nucleoids; extensively damaged cells with dot-likes nucleoids stained with SYTOX green (b) and bottom layer cells with undamaged plasma membranes, resulting SYTOX-Green negative (b arrow) Bar = 5 μm
When biofilms of strain CCMEE 057 exposed to 10 kJ/m2 of UVC were stained with the cell- impermeant nucleic acid dye SYTOX-Green the upper cell layers had damaged plasma membranes and green-fluorescent nucleoids (Fig. 4b). Extensively damaged cells released SYTOX-Green stained nucleoids or exhibited fragmented, dot-like nucleoids in the cytoplasm (Fig. 4b). SYTOX- Green negative cells occurred at the bottom cell layers of biofilms (Fig. 4b, arrow). A positive colony-forming ability was scored for each UVC-exposed sample.
EPS Composition of Biofilms and Planktonic Cells
When the EPS composition was assessed by CLSM imaging combined with fluorescent probes, differences were revealed between biofilms and planktonic samples. In strain CCMEE 057 the staining with the lectin ConA revealed no differences in the abundance of α-mannopyranosyl and α- gluocopyranyl residues in the EPS of biofilms and planktonic cells (Fig. 5a–b), while the lipophilic green fluorescent BODIPY revealed abundant lipidic compounds in biofilms (Fig. 5d) in contrast to planktonic cells (Fig. 5c).
CLSM images of stained EPS in biofilms and planktonic cells of Chroococcidiopsis sp. CCMEE 057. Polysaccharides with α-mannopyranosyl and α-glucopyranosyl revealed by the fluorescence-labelled lectin ConA in biofilms (a) and planktonic cells (b); lipids stained highlighted by BODIPY FL C12 in biofilms (c) and planktonic cells (d); Scale bar = 10 μm
In strain CCMEE 029, EPS of biofilms and planktonic samples exhibited fluorescence after staining with ConA; while the BODYPY staining indicated an increase in lipids in biofilms unlike in planktonic cells (not shown).
Discussion
When Chroococcidiopsis sp. strain CCMEE 057 from the Sinai Desert and strain CCMEE 029 from the Negev Desert were exposed to space and Martian simulations in the dried status either as biofilms or multilayered planktonic samples, survival was better supported by the biofilm lifestyle. Biofilms provided a protective environment for Chroococcidiopsis cells as suggested by the quantification and visualization at the single-cell level of the induced damage. PMA-qPCR assay quantified cells with undamaged plasma membranes by amplifying a target gene, as opposed to damaged cells where the influx of the DNA-binding PMA dye impairs the amplification (Nocker and Camper 2008). DNA damage was evaluated by using PCR-stop assays: real-time qPCR quantified DNA damage via a target gene amplification (Fajardo-Cavazos et al. 2010), whereas RAPD revealed DNA damage by yielding altered PCR fingerprints (Trombert et al. 2007). Finally, the induced damage was visualized by CLSM imaging of biofilms and planktonic samples. Since exposure to unattenuated polychromatic UV radiation suggested that Chroococcidiopsis biofilms of both the investigated strains did not survive doses higher than 1.5 × 103 kJ/m2 (not shown), samples were exposed to space vacuum or Martian atmosphere of 780 Pa pressure under UV radiation of 5 × 105 kJ/m2 (with a 0.1 % ND filter). After exposure to space and Martian simulations, biofilms of strain CCMEE 057 showed reduced damage to plasma membranes and genomic DNA compared to planktonic multilayers. Furthermore, unlike biofilms, planktonic samples after space simulations lost their colony-forming ability, thus suggesting a failure in repairing the accumulated damage. Biofilms of strain CCMEE 029 were less tolerant than those of strain CCMEE 057, being able to withstand only vacuum and Martian atmosphere and pressure; nevertheless under these conditions biofilms were more resistant than their planktonic counterparts. Indeed, unlike planktonic samples, biofilms of strain CCMEE 029 survived 1.5 × 103 kJ/m2 of unattenuated polychromatic UV radiation (not shown). After space simulation CLSM imaging evealed extensively bleached photosynthetic pigments in the top cell layers of planktonic samples compared to biofilms; also PCR-based assays indicated that planktonic cells were more susceptible to damage than biofilms were.
Our findings contribute to the appreciation of the survival potential of Chroococcidiopsis under space and Martian conditions and offered new insights into the endurance of phototrophic biofilm. Previouly dried monolayers and multilayers of Chroococcidiopsis exposed to ground-based simulations or Low Earth Orbit were obtained from planktonic cultures subjected to air-drying. Cells of strain CCMEE 029, when augmented with other cyanobacteria at the top of an epilithic community survived 584 days in space, where they experienced an extraterrestrial ultraviolet dose of 5.15 × 105 kJ/m2. However, since monolayers of strain CCMEE 029 tolerated up to 30 kJ/m2 of simulated Mars flux (Cockell et al. 2005) and 1 day of UV fluxes in the Atacama Desert (Cockell et al. 2008), it was suggested that Chroococcidiopsis survival in space benefited from the shielding provided by surface layers (Cockell et al. 2011).
In the present work, the protective role played by top cell layers was revealed by CLSM imaging and PCR-stop assay: Damaged cells were visualized in the first 3–4 cell layers of UVC-exposed biofilms but no significant increase in DNA damage occurred after 10 J/m2 (up to 10 kJ/m2). A protective role was reported for a triple layer of B. subtilis spores: Survival was reduced by 52 kJ/m2 of UV radiation, but it was not further reduced by higher doses (Mancinelli and Klovstad 2000).
The endurance of Chroococcidiopsis biofilms compared to planktonic cells is in line with the advantage of living as a biofilm when facing extreme conditions. However, while the planktonic- biofilm transition has been extensively investigated in bacteria (Stanley and Lazazzera 2004), in the case of cyanobacteria it remains largely unknown.
Since EPS are a key component in the biofilm endurance under stress, the EPS composition in Chroococcidiopsis biofilms and planktonic cells was investigated by using CLSM combined with molecular probes. The staining with the fluorescence-labelled lectin ConA revealed no differences in polysaccharides containing α-mannopyranosyl and α-glucopyranosyl in biofilms and planktonic samples. Whereas the lipophilic fluorophore BODIPY identified lipid accumulation in biofilms as opposed to planktonic cells. Although lipids have been stained in bacterial biofilms by using nile red (Lawrence et al. 2003), BODIPY is more suitable for lipid staining (Govender et al. 2012) and used to stain outer cell layers and photosynthetic membranes in cyanobacteria (Mullineaux et al. 2008). Abundant polysaccharides and lipids have been reported in the matrix encapsulating Chroococcidiopsis cells in desert stones and laboratory-dried samples (Grilli Caiola et al. 1996).
The EPS involvement in UV tolerance by scavenging reactive oxygen species, by increasing the path length of the incident radiation and by providing a matrix for UV-absorbing pigments, carotenoids and enzymes has been reported (Chen et al. 2009; Baqué et al. 2013; Helm and Potts 2012). Nevertheless, the composition and biosynthetic pathways of EPS in cyanobacteria are not well understood (Pereira et al. 2009). Once the EPS composition of Chroococcidiopsis biofilms and planktonic cells is more deeply characterized, insight will be gained into the biofilm endurance under space and Martian conditions. Furthermore the comparison of the EPS composition in biofilms and planktonic cells might contribute explaining the biofilm tolerance to air-drying; in fact, the real-time qPCR assay revealed an increase in DNA damage upon drying in planktonic cells compared to biofilms.
In conclusion, our results suggest that biofilms consisting of about 10 cell layers might support the endurance of Chroococcidiopsis (CCMEE 057) for 1 year in space, under 0.1 % solar irradiance as expected during the EXPOSE-R2 mission. Layers of B. subtilis with a thickness of 5–10 spores, after 1.5 years exposure in space under full solar radiation exhibited 3-fold reduced survival, while a 1-fold reduction occurred under 0.1 % solar radiation (Moeller et al. 2012). In the present work, given that the total UV polychromatic dose experienced by Chroococcidiopsis biofilms was of about 5 × 102 kJ/m2 (due the ND filter) and that the maximum UV flux on a clear day on the Mars equator (in the 200–400 nm range) is about 50 W/m2 (Schuerger et al. 2003), an approximate survival of about 2 h and 45 min was extrapolated for Chroococcidiopsis biofilms on the Mars surface. It now remains to investigate the resistance under space and Martian simulations of Chroococcidiopsis biofilms grown in the presence of Martian minerals. Indeed, in the frame of the BIOMEX project, dried cells from a planktonic culture of strain CCMEE 029 mixed with different Martian and lunar mineral analogues, survived up to 1.5 × 103 kJ/m2 of UV polychromatic radiation in contrast to controls in absence of minerals (Baqué, personal communication). The future study which we propose might also contribute to unravelling the influence of minerals on the EPS production and role in the evolution of phototrophic biofilms on early Earth when the UV radiation climate was similar to that of Mars.
Our results further support the relevance of ground-based simulations to unravel how biological compounds and organisms thrive in, or are modified by, the space conditions in preparation for their exposure outside the ISS. New generation astrobiological experiments are being carried on board of cubesats that orbit where the radiation dose is significantly higher (at least one order of magnitude) than on ISS. Given that cubesats stay in orbit for several months, using in situ measurement technology they make it possible to study the dynamics of space-induced changes (i.e. gravitational effects) so extending researches previously limited to before-and-after-flight comparisons (Woellert et al. 2011).
References
Baqué M, Viaggiu E, Scalzi G, Billi D (2013) Endurance of the endolithic desert cyanobacterium Chroococcidiopsis under UVC radiation. Extremophiles 17:161–169. doi:10.1007/s00792-012-0505-5
Billi D (2009) Subcellular integrities in Chroococcidiopsis sp. CCMEE 029 survivors after prolonged desiccation revealed by molecular probes and genome stability assays. Extremophiles 13:49–57. doi:10.1007/s00792-008-0196-0
Billi D (2012) Anhydrobiotic rock-inhabiting cyanobacteria: potential for astrobiology and biotechnology. In: Stan-Lotter H, Fendrihan F (eds) Adaptation of microbial life organisms in extreme environments: research and application. Springer Wien, New York, pp 119–132
Billi D, Grilli Caiola M, Paolozzi L, Ghelardini P (1998) A method for DNA extraction from the desert cyanobacterium Chroococcidiopsis and its application to identification of ftsZ. Appl Environ Microbiol 64:4053–4056
Billi D, Friedmann EI, Hofer KG, Grilli Caiola M, Ocampo-Friedmann R (2000) Ionizing-radiation resistance in the desiccation-tolerant cyanobacterium Chroococcidiopsis. Appl Environ Microbiol 66:1489–1492
Billi D, Viaggiu E, Cockell CS, Rabbow E, Horneck G, Onofri S (2011) Damage escape and repair in dried Chroococcidiopsis spp. from hot and cold deserts exposed to simulated space and Martian conditions. Astrobiology 11:65–73. doi:10.1089/ast.2009.0430
Chen LZ, Wang GH, Hong S, Liu A, Li C, Liu YD (2009) UV-B-induced oxidative damage and protective role of exopolysaccharides in desert cyanobacterium Microcoleus vaginatus. J Integr Plant Biol 51:194–200
Cockell CS, Schuerger AC, Billi D, Friedmann EI, Panitz C (2005) Effects of a simulated martian UV flux on the cyanobacterium, Chroococcidiopsis sp. 029. Astrobiology 5:127–140. doi:10.1089/ast.2005.5.127
Cockell CS, McKay CP, Warren-Rhodes K, Horneck G (2008) Ultraviolet radiation-induced limitation to epilithic microbial growth in arid deserts—dosimetric experiments in the hyperarid core of the Atacama Desert. J Photochem Photobiol B 90:79–87. doi:10.1016/j.jphotobiol.2007.11.009
Cockell CS, Rettberg P, Rabbow E, Olsson-Francis K (2011) Exposure of phototrophs to 548 days in low Earth orbit: microbial selection pressures in outer space and on early earth. ISME J :1–12. doi:10.1038/ismej.2011.46
de Vera J-P, Boettger U, de la Torre Noetzel R, Sánchez FJ, Grunow D, Schmitz N, Lange C, Hübers H-W, Billi D, Baqué M, Rettberg R, Rabbow E, Reitz G, Berger T, Möller R, Bohmeier M, Horneck G, Westall F, Jänchen J, Fritz J, Meyer C, Onofri S, Selbmann L, Zucconi L, Kozyrovska N, Leya T, Foing B, Demets, Cockell CS, Bryce C, Wagner D, Serrano P, Edwards HGM, Joshi J, Huwe B, Ehrenfreund P, Ott S, Meessen J, Feyh N, Szewzyk U, Jaumann R, Spohn T (2012) Supporting Mars exploration: BIOMEX in low Earth orbit and further astrobiological studies on the Moon using raman and PanCam technology. Planet Space Sci 74:103–110. doi:10.1016/j.pss.2012.06.010
Fajardo-Carvazos P, Schuerger AC, Nicholson WL (2010) Exposure of DNA and Bacillus subtilis spores to simulated Martian environments: use of Quantitative PCR (qPCR) to measure inactivation rates of DNA to function as a template molecule. Astrobiology 10:403–411. doi:10.1089/ast.2009.0408
Flemming HC, Wingender J (2010) The biofilm matrix. Nat Rev Microbiol 8:623–633. doi:10.1038/nrmicro2415
Friedmann EI (1980) Endolithic microbial life in hot and cold deserts. Orig Life Evol Biosph 10:223–235. doi:10.1007/BF00928400
Govender T, Ramanna L, Rawat I, Bux F (2012) BODIPY staining, an alternative to the Nile Red fluorescence method for the evaluation of intracellular lipids in microalgae. Bioresour Technol 114:507–511. doi:10.1016/j.biortech.2012.03.024
Grilli Caiola M, Billi D, Friedmann EI (1996) Effect of desiccation on envelopes of the cyanobacterium Chroococcidiopsis sp. (Chroococcales). Eur J Phycol 31:97–105
Helm RF, Potts M (2012) Extracellular Matrix (ECM). In: Whitton BA (ed) Ecology of 443 cyanobacteria II. Springer, Netherlands, pp 461–480
Horneck G, Becker H, Reitz G (1994) Long-term survival of bacterial spores in space. Adv Space Res 14:41–45
Horneck G, Klaus DM, Mancinelli RL (2010) Space microbiology. Microbiol Mol Biol Rev 74:121–156
Lawrence JR, Swerhone GD, Leppard GG, Araki T, Zhang X, West MM, Hitchcock AP (2003) Scanning transmission X-ray, laser scanning, and transmission electron microscopy mapping of the exopolymeric matrix of microbial biofilms. Appl Environ Microbiol 69:5543–5554. doi:10.1128/AEM.69.9.5543-5554.2003
Mancinelli RL, Klovstad M (2000) Martian soil and UV radiation: microbial viability assessment on spacecraft surfaces. Planet Space Sci 48:1093–1097. doi:10.1016/S0032-0633(00)00083-0
Moeller R, Reitz G, Nicholson, The Protect Team WL, Team WL, Horneck G (2012) Mutagenesis in bacterial spores exposed to space and simulated martian conditions: data from the EXPOSE-E spaceflight experiment PROTECT. Astrobiology 12:457–468. doi:10.1089/ast.2011.0739
Mullineaux CW, Mariscal V, Nenninger A, Khanum H, Herrero A, Flores E, Adams DG (2008) Mechanism of intercellular molecular exchange in heterocyst-forming cyanobacteria. EMBO J 27:1299–1308. doi:10.1038/emboj.2008.66
Nocker A, Camper AK (2008) Novel approaches toward preferential detection of viable cells using nucleic acid amplification techniques. FEMS Microbiol Lett 291:137–142. doi:10.1111/j.1574-6968.2008.01429.x
Onofri S, de la Torre R, de Vera JP, Ott S, Zucconi L, Selbmann L, Scalzi G, Venkateswaran KJ, Rabbow E, Sánchez Iñigo FJ, Horneck G (2012) Survival of rock-colonizing organisms after 1.5 years in outer space. Astrobiology 12:508–516. doi:10.1089/ast.2011.0736
Pereira S, Zille A, Micheletti E, Moradas-Ferreira P, De Philippis R, Tamagnini P (2009) Complexity of cyanobacterial exopolysaccharides: composition, structures, inducing factors and putative genes involved in their biosynthesis and assembly. FEMS Microbiol Rev 33:917–941
Poli A, Anzelmo G, Nicolaus B (2010) Bacterial exopolysaccharides from extreme marine habitats: production, characterization and biological activities. Mar Drugs 8:1779–1802. doi:10.3390/md8061779
Rabbow E, Horneck G, Rettberg P, Schott J-U, Panitz C, L’Afflitto A, von Heise-Rotenburg R, Willnecker R, Baglioni P, Hatton J, Dettmann J, Demets R, Reitz G (2009) EXPOSE, an astrobiological exposure facility on the international space station—from proposal to flight. Orig Life Evol Biosph 39:581–598. doi:10.1007/s11084-009-9173-6
Rabbow E, Rettberg P, Barczyk S, Bohmeier M, Parpart A, Panitz C, Horneck G, von Heise-Rotenburg R, Hoppenbrouwers T, Willnecker R, Baglioni P, Demets R, Dettmann J, Reitz G (2012) EXPOSE-E: an ESA astrobiology mission 1.5 years in space. Astrobiology 12:374–386. doi:10.1089/ast.2011.0760
Sancho LG, de la Torre R, Horneck G, Ascaso C, de los Rios A, Pintado A, Wierzchos JM, Schuster M (2007) Lichens survive in space: results from the 2005 LICHENS experiment. Astrobiology 7:443–454. doi:10.1089/ast.2006.0046
Schuerger AC, Mancinelli RL, Kern RG, Rothschild LJ, McKay CP (2003) Survival of Bacillus subtilis on spacecraft surfaces under simulated Martian environments: implications for the forward contamination of Mars. Icarus 165:253–276. doi:10.1016/S0019-1035(03)00200-8
Stanley NR, Lazazzera BA (2004) Environmental signals and regulatory pathways that influence biofilm formation. Mol Microbiol 52:917–924. doi:10.1111/j.1365-2958.2004.04036.x
Stivaletta N, Barbieri R, Billi D (2012) Microbial colonization of the salt desposits in the driest place in Atacama Desert (Chile). Orig Life Evol Biosph 42:187–200. doi:10.1007/s11084-012-9289-y
Tromber A, Irazoqui H, Martin C, Zalazar F (2007) Evaluation of UV-C induced changes in Escherichia coli using repetitice extragenic palindromic-polymerase chain reaction (REP-PCR). J Photochem Photobiol B 89:44–49. doi:10.1016/j.jphotobiol.2007.08.003
Wierzchos J, Ascaso C, Mckay CP (2006) Endolithic cyanobacteria in halite rocks from the hyperarid core of the Atacama Desert. Astrobiology 6:1–7. doi:10.1089/ast.2006.6.415
Woellert K, Ehrenfreund P, Ricco AJ, Hertzfeld H (2011) Cost-effective science and technology platforms for emerging and developing nations. Adv Space Resx 47:663–684. doi:10.1016/j.asr.2010.10.009
Xu J, Campbell JM, Zhang N, Hickey WJ, Sahai N (2012) Did mineral surface chemistry and toxicity contribute to evolution of microbial extracellular polymeric substances? Astrobiology 12:785–798. doi:10.1089/ast.2011.0776
Acknowledgments
This research was supported by the Italian Space Agency (ASI) and partially supported by the Italian Ministry of Foreign Affair. The authors thank Dr. Elena Romano, Centre of Advanced Microscopy “P. B. Albertano”, University of Rome “Tor Vergata”, for her skillful assistance in using the facility.
Author information
Authors and Affiliations
Corresponding author
Additional information
Mickael Baqué and Giuliano Scalzi contributed equally to this work.
This manuscript is in memoriam of Imre E. Fridamm (1921–2007) and Roseli Ocampo-Friedamm (1937–2005)
Paper presented at the 12th European Workshop on Astrobiology "EANA'12" in Stockholm, Sweden (October 15 to 17, 2012). Editors Axel Brandenburg and Nils Holm
Rights and permissions
About this article
Cite this article
Baqué, M., Scalzi, G., Rabbow, E. et al. Biofilm and Planktonic Lifestyles Differently Support the Resistance of the Desert Cyanobacterium Chroococcidiopsis Under Space and Martian Simulations. Orig Life Evol Biosph 43, 377–389 (2013). https://doi.org/10.1007/s11084-013-9341-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11084-013-9341-6