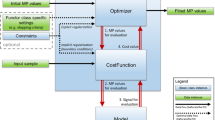
Abstract
PDE-constrained optimization problems find many applications in medical image analysis, for example, neuroimaging, cardiovascular imaging, and oncologic imaging. We review the related literature and give examples of the formulation, discretization, and numerical solution of PDE-constrained optimization problems for medical imaging. We discuss three examples. The first is image registration, the second is data assimilation for brain tumor patients, and the third is data assimilation in cardiovascular imaging. The image registration problem is a classical task in medical image analysis and seeks to find pointwise correspondences between two or more images. Data assimilation problems use a PDE-constrained formulation to link a biophysical model to patient-specific data obtained from medical images. The associated optimality systems turn out to be sets of nonlinear, multicomponent PDEs that are challenging to solve in an efficient way. The ultimate goal of our work is the design of inversion methods that integrate complementary data, and rigorously follow mathematical and physical principles, in an attempt to support clinical decision making. This requires reliable, high-fidelity algorithms with a short time-to-solution. This task is complicated by model and data uncertainties, and by the fact that PDE-constrained optimization problems are ill-posed in nature, and in general yield high-dimensional, severely ill-conditioned systems after discretization. These features make regularization, effective preconditioners, and iterative solvers that, in many cases, have to be implemented on distributed-memory architectures to be practical, a prerequisite. We showcase state-of-the-art techniques in scientific computing to tackle these challenges.









Similar content being viewed by others
Notes
In general, constraints can either be hard or soft. Hard constraints are a set of conditions that the variables are required to satisfy. Soft constraints are penalties for variables that appear in the objective functional; they penalize deviation of the variables from a condition.
We note that high-resolution imaging technologies have emerged; an example is CLARITY imaging (Chung and Deisseroth 2013; Tomer et al. 2014; Kutten et al. 2017); we revisit this ex-situ imaging technique briefly in Sect. 2. High-resolution imaging techniques are not yet available in a clinical setting. Resolution levels for routinely collected imaging data are between 0.5 and 5 mm in each spatial direction (depending on the imaging modality).
Similar formulations can be found in other applications, e.g., geophysical sciences Fohring et al. (2014).
The number of unknowns is the number of discretization points in space times the number of discretization points in time for the velocity times the dimensionality of the ambient space (we invert for a time-dependent vector field).
A stationary velocity field v is a velocity field that is constant in time, as opposed to a nonstationary—i.e., time-dependent or transient—velocity field. We note that stationary velocity fields do not cover the entire space of diffeomorphisms and do not provide a Riemannian metric on this space, something that may be desirable in certain applications (Beg et al. 2005; Miller 2004; Zhang and Fletcher 2015); this requires time-dependent velocities. The work in Mang and Biros (2015) uses a Galerkin method to control the number of unknowns in time. It is shown experimentally that stationary and nonstationary velocities yield an equivalent registration quality in terms of data mismatch.
The resolution of these datasets can reach \(5 \upmu m\times 5 \upmu m\times 5 \upmu m\) resulting in \({\mathcal {O}}(4.8\hbox { TB})\) of data (if stored with half precision) Tomer et al. (2014). Overall, this results in 2.4 trillion discretization points in space.
Other distance measures, such as mutual information, normalized gradient fields, or cross correlation can be used (see Modersitzki 2004, 2009 for an overview). In our formulation, changing the distance measure will affect the first term in the objective functional and the terminal condition of the adjoint equation (8) (see Sect. 5.1.1).
The white matter fibre architecture can be estimated from data measured by so-called diffusion tensor imaging, a magnetic resonance imaging technique that measures the anisotropy of diffusion in the human brain. The result of this measurement is a tensor field \(\tilde{k}\) that can directly be inserted into (4).
Mutual information is a statistical distance measure that originates from information theory. As opposed to the squared \(L^2\) distance used in the present work (see Sect. 2), which is used for registering images acquired with the same modality, mutual information is used for the registration of images that are acquired with different modalities (e.g., the registration of computed tomography and magnetic resonance images). Mutual information assess the statistical dependence between two random variables (in our case image intensities; see Modersitzki 2004, 2009; Sotiras et al. 2013 for more details).
We neglect the periodic boundary conditions for simplicity.
Instead of solving an advection or continuity equation as done in the Eulerian formulation we present here, it has been suggested to solve for the state and adjoint variables using the diffeomorphism y (which involves interpolation operations instead of the solution of a transport equation) Vialard et al. (2012).
We neglect the Neumann boundary conditions for simplicity.
The data assimilation problem in Sect. 3 requires Neumann boundary conditions on \(\partial {\varOmega }_B\), with \({\varOmega }_B\subset {\varOmega }\). We use a penalty approach to approximate these boundary conditions. We apply periodic boundary conditions on \(\partial {\varOmega }\) and set the reaction and diffusion coefficients in (4a) to sufficiently small penalty parameters \(k^\epsilon \rightarrow 0\) and \(\rho ^\epsilon \rightarrow 0\) outside of \({\varOmega }_B\); see, e.g., Gholami et al. (2016), Hogea et al. (2008b), Mang (2014) and Mang et al. (2012).
Note that the \(\inf\)-\(\sup\) condition for pressure spaces arising in finite element discretizations of Stokes problems (Brezzi and Fortin 1991, p. 200ff.) is not an issue with our scheme (incompressibility constraint in diffeomorphic registration problem).
The order \(\tilde{n}\) of the optimality system depends on the problem. For the diffeomorphic registration case the control variable \({\mathbf {w}}\) is given by the velocity field \({\mathbf {v}}\in {\mathbf {R}}^{dn}\), i.e., \(\tilde{n}\equiv dn\); for the tumor case \({\mathbf {w}}\) is given by \(p\in {\mathbf {R}}^{n_p}\), i.e., \(\tilde{n}\equiv n_p\).
Our implementation also features a trust region method.
Additional information on the data sets, the imaging protocol, and the preprocessing can be found in Christensen et al. (2006) and at http://www.nirep.org/.
References
Akcelik V, Biros G, Ghattas O (2002) Parallel multiscale Gauss–Newton–Krylov methods for inverse wave propagation. In: Proceeding of the ACM/IEEE conference on supercomputing, pp 1–15
Akcelik V, Biros G, Ghattas O, Hill J, Keyes D, van Bloemen Wanders B (2006) Parallel algorithms for PDE constrained optimization. In: Parallel processing for scientific computing, vol 20, chap. 16. SIAM, Philadelphia, pp 291–322
Alexanderian A, Petra N, Stadler G, Ghattas O (2016) A fast and scalable method for A-optimal design of experiments for infinite-dimensional Bayesian nonlinear inverse problems. SIAM J Sci Comput 38(1):A243–A272
Amit Y (1994) A nonlinear variational problem for image matching. SIAM J Sci Comput 15(1):207–224
Andreev R, Scherzer O, Zulehner W (2015) Simultaneous optical flow and source estimation: space-time discretization and preconditioning. Appl Numer Math 96:72–81
Angenent S, Haker S, Tannenbaum A (2003) Minimizing flows for the Monge-Kantrovich problem. SIAM J Math Anal 35(1):61–97
Arridge SR (1999) Optical tomography in medical imaging. Inverse Probl 15:R41–R93
Arridge SR, Schotland JC (2009) Optical tomography: forward and inverse problems. Inverse Probl 25(12):123,010
Arsigny V, Commowick O, Pennec X, Ayache N (2006) A Log-Euclidean framework for statistics on diffeomorphisms. Proceedings of the medical image computing and computer-assisted intervention, vol LNCS 4190:924–931
Ashburner J (2007) A fast diffeomorphic image registration algorithm. NeuroImage 38(1):95–113
Ashburner J, Friston KJ (2011) Diffeomorphic registration using geodesic shooting and Gauss-Newton optimisation. NeuroImage 55(3):954–967
Atuegwu NC, Colvin DC, Loveless ME, Xu L, Gore JC, Yankeelov TE (2012) Incorporation of diffusion-weighted magnetic resonance imaging data into a simple mathematical model of tumor growth. Phys Med Biol 57(1):225
Avants B, Schoenemann PT, Gee JC (2006) Lagrangian frame diffeomorphic image registration: morphometric comparison of human and chimpanzee cortex. Med Image Anal 10:397–412
Avants BB, Epstein CL, Brossman M, Gee JC (2008) Symmetric diffeomorphic image registration with cross-correlation: evaluating automated labeling of elderly and neurodegenerative brain. Med Image Anal 12(1):26–41
Avants BB, Tustison NJ, Song G, Cook PA, Klein A, Gee JC (2011) A reproducible evaluation of ANTs similarity metric performance in brain image registration. NeuroImage 54:2033–2044
Axel L (2002) Biomechanical dynamics of the heart with MRI. Annu Rev Biomed Eng 4:321–347
Balay S, Abhyankar S, Adams MF, Brown J, Brune P, Buschelman K, Eijkhout V, Gropp WD, Kaushik D, Knepley MG, McInnes LC, Rupp K, Smith BF, Zhang H (2016) PETSc users manual. Tech. Rep. ANL-95/11-Revision 3.7. Argonne National Laboratory
Barbu V, Marinoschi G (2016) An optimal control approach to the optical flow problem. Syst Control Lett 87:1–9
Barthel W, John C, Tröltsch F (2010) Optimal boundary control of a system of reaction diffusion equations. Z Angew Math Mech 90:966–982
Beg MF, Miller MI, Trouvé A, Younes L (2005) Computing large deformation metric mappings via geodesic flows of diffeomorphisms. Int J Comput Vis 61(2):139–157
Bellomo N, Li NK, Maini PK (2008) On the foundations of cancer modelling: selected topics, speculations, and perspectives. Math Models Methods Appl Sci 18(4):593–646
Benamou JD, Brenier Y (2000) A computational fluid mechanics solution to the Monge-Kantorovich mass transfer problem. Numer Math 84:375–393
Benzi M, Golub GH, Liesen J (2005) Numerical solution of saddle point problems. Acta Numer 14:1–137
Benzi M, Haber E, Taralli L (2009) Multilevel algorithms for large-scale interior point methods. SIAM J Sci Comput 31(6):4152–4175
Benzi M, Haber E, Taralli L (2011) A preconditioning technique for a class of PDE-constrained optimization problems. Adv Comput Math 35(2–4):149–173
Biegler LT, Ghattas O, Heinkenschloss M, van Bloemen Waanders B (2003) Large-scale PDE-constrained optimization. Springer, Berlin
Biegler LT, Ghattas O, Heinkenschloss M, Keyes D, van Bloemen Waanders B (2007) Real-time PDE-constrained optimization. SIAM, Philadelphia
Biros G, Ghattas O (1999) Parallel Newton–Krylov methods for PDE-constrained optimization. In: Proceedings of the ACM/IEEE conference on supercomputing, pp 28–40
Biros G, Ghattas O (2005a) Parallel Lagrange–Newton–Krylov–Schur methods for PDE-constrained optimization-part I: the Krylov-Schur solver. SIAM J Sci Comput 27(2):687–713
Biros G, Ghattas O (2005b) Parallel Lagrange–Newton–Krylov–Schur methods for PDE-constrained optimization-part II: the Lagrange-Newton solver and its application to optimal control of steady viscous flows. SIAM J Sci Comput 27(2):714–739
Bistoquet A, Parks WJ, Skrinjar O (2006) Myocardial deformation recovery using a 3D biventricular incompressible model. In: Proceedings of the biomedical image registration (Lecture Notes in computer science), vol 4057. Springer, Berlin, pp 110–119
Bluemke DA, Krupinski EA, Ovitt T, Gear K, Unger E, Axel L, Boxt LM, Casolo G, Ferrari VA, Funaki B, Globits S, Higgins CB, Julsrud P, Lipton M, Mawson J, Nygren A, Pennell DJ, Stillman A, White RD, Wichter T, Marcus F (2003) MR imaging of arrhythmogenic right ventricular cardiomyopathy: morphologic findings and interobserver reliability. Cardiology 99(3):153–162
Borzì A, Ito K, Kunisch K (2002) Optimal control formulation for determining optical flow. SIAM J Sci Comput 24(3):818–847
Borzì A, Schulz V (2012) Computational optimization of systems governed by partial differential equations. SIAM, Philadelphia
Brezzi F, Fortin M (eds) (1991) Mixed and hybrid finite element methods. Springer, Berlin
Burger M, Modersitzki J, Ruthotto L (2013) A hyperelastic regularization energy for image registration. SIAM J Sci Comput 35(1):B132–B148
Castillo E, Lima JAC, Bluemke DA (2003) Regional myocardial function: advances in MR imaging and analysis. Radiographics 23:S127–S140
Chen K (2011) Optimal control based image sequence interpolation. Ph.D. thesis, University of Bremen
Chen K, Lorenz DA (2011) Image sequence interpolation using optimal control. J Math Imaging Vis 41:222–238
Chen X, Summers RM, Yoa J (2012) Kidney tumor growth prediction by coupling reaction-diffusion and biomechanical model. IEEE Trans Biomed Eng 60(1):169–173
Christensen GE, Rabbitt RD, Miller MI (1994) 3D brain mapping using a deformable neuroanatomy. Phys Med Biol 39(3):609–618
Christensen GE, Rabbitt RD, Miller MI (1996) Deformable templates using large deformation kinematics. IEEE Trans Image Process 5(10):1435–1447
Christensen GE, Geng X, Kuhl JG, Bruss J, Grabowski TJ, Pirwani IA, Vannier MW, Allen JS, Damasio H (2006) Introduction to the non-rigid image registration evaluation project. Proceedings of the biomedical image registration, vol LNCS 4057:128–135
Chung K, Deisseroth K (2013) CLARITY for mapping the nverous system. Nat Methods 10:508–513
Clatz O, Sermesant M, Bondiau PY, Delingette H, Warfield SK, Malandain G, Ayache N (2005) Realistic simulation of the 3D growth of brain tumors in MR images coupling diffusion with biomechanical deformation. IEEE Trans Med Imaging 24(10):1334–1346
Cocosco C, Kollokian V, Kwan RKS, Evans AC (1997) Brainweb: online interface to a 3D MRI simulated brain database. NeuroImage 5:425
Colin T, Iollo A, Lagaert JB, Saut O (2014) An inverse problem for the recovery of the vascularization of a tumor. J Inverse Ill Posed Probl 22(6):759–786
Collis J, Connor AJ, Paczkowski M, Kannan P, Pitt-Francis J, Byrne HM, Hubbard ME (2017) Bayesian calibration, validation and uncertainty quantification for predictive modelling of tumour growth: a tutorial. Bull Math Biol 79(4):939–974
Crippa G (2007) The flow associated to weakly differentiable vector fields. Ph.D. thesis, University of Zürich
Croft W, Elliott CM, Ladds G, Stinner B, Venkataraman C, Weston C (2015) Parameter identification problems in the modelling of cell motility. J Math Biol 71(2):399–436
Czechowski K, Battaglino C, McClanahan C, Iyer K, Yeung PK, Vuduc R (2012) On the communication complexity of 3D FFTs and its implications for exascale. In: Proceedings of the ACM/IEEE Conference on supercomputing, pp 205–214
Delingette H, Billet F, Wong KCL, Sermesant M, Rhode K, Ginks M, Rinaldi C, Razavi R, Ayache N (2012) Personalization of cardiac motion and contractility from images using variational data assimilation. IEEE Trans Biomed Eng 59(1):20–24
Dembo RS, Steihaug T (1983) Truncated-Newton algorithms for large-scale unconstrained optimization. Math Program 26(2):190–212
Dontchev AL, Hager WW, Veliov VM (2000) Second-order Runge-kutta approximations in control constrained optimal control. SIAM J Numer Anal 38(1):202–226
Dupuis P, Gernander U, Miller MI (1998) Variational problems on flows of diffeomorphisms for image matching. Q Appl Math 56(3):587–600
Eisentat SC, Walker HF (1996) Choosing the forcing terms in an inexact Newton method. SIAM J Sci Comput 17(1):16–32
Eklund A, Dufort P, Forsberg D, LaConte SM (2013) Medical image processing on the GPU-past, present and future. Med Image Anal 17(8):1073–1094
Ellingwood ND, Yin Y, Smith M, Lin CL (2016) Efficient methods for implementation of multi-level nonrigid mass-preserving image registration on GPUs and multi-threaded CPUs. Comput Methods Program Biomed 127:290–300
Engl H, Hanke M, Neubauer A (1996) Regularization of inverse problems. Kluwer Academic Publishers, Dordrecht
Falcone M, Ferretti R (1998) Convergence analysis for a class of high-order semi-Lagrangian advection schemes. SIAM J Numer Anal 35(3):909–940
Figueiredo IN, Leal C (2013) Physiologic parameter estimation using inverse problems. SIAM J Appl Math 73(3):1164–1182
Figueiredo IN, Figueiredo PN, Almeida N (2011) Image-driven parameter estimation in absorption-diffusion models of chromoscopy. SIAM J Imaging Sci 4(3):884–904
Fischer B, Modersitzki J (2008) Ill-posed medicine–an introduction to image registration. Inverse Probl 24(3):1–16
Fluck O, Vetter C, Wein W, Kamen A, Preim B, Westermann R (2011) A survey of medical image registration on graphics hardware. Comput Methods Programs Biomed 104(3):e45–e57
Fohring J, Haber E, Ruthotto L (2014) Geophysical imaging for fluid flow in porous media. SIAM J Sci Comput 36(5):S218–S236
Garcke H, Lam KF, Sitka E, Styles V (2016) A Cahn-Hilliard-Darcy model for tumour growth with chemotaxis and active transport. Math Methods Appl Sci 26(6):1095–1148
Geweke J, Tanizaki H (1999) On Markov chain Monte Carlo methods for nonlinear and non-Gaussian state-space models. Commun Stat Simul Comput 28(4):867–894
Geweke J, Tanizaki H (2003) Note on the sampling distribution for the Metropolis-Hastings algorithm. Commun Stat Theory Methods 32(4):2003
Gholami A, Hill J, Malhotra D, Biros G (2016a) AccFFT: a library for distributed-memory FFT on CPU and GPU architectures. In review (arXiv:1506.07933)
Gholami A, Mang A, Biros G (2016b) An inverse problem formulation for parameter estimation of a reaction-diffusion model of low grade gliomas. J Math Biol 72(1):409–433. https://doi.org/10.1007/s00285-015-0888-x
Gholami A, Mang A, Scheufele K, Davatzikos C, Mehl M, Biros G (2017) A framework for scalable biophysics-based image analysis. Proc ACM/IEEE Conf Supercomput 19:1–13. https://doi.org/10.1145/3126908.3126930
Goenezen S, Dord JF, Sink Z, Barbone PE, Jiang J, Hall TJ, Oberai AA (2012) Linear and nonlinear elastic modulus imaging: an application to breast cancer diagnosis. IEEE Trans Med Imaging 31(8):1628–1637
Gooya A, Pohl KM, Bilello M, Cirillo L, Biros G, Melhem ER, Davatzikos C (2013) GLISTR: glioma image segmentation and registration. IEEE Trans Med Imaging 31(10):1941–1954
Grama A, Gupta A, Karypis G, Kumar V (2003) An Introduction to parallel computing: design and analysis of algorithms, 2nd edn. Addison Wesley, Boston
Griewank A (1992) Achieving logarithmic growth of temporal and spatial complexity in teverse automatic differentiation. Optim Methods Softw 1:35–54
Gu X, Pand H, Liang Y, Castillo R, Yang D, Choi D, Castillo E, Majumdar A, Guerrero T, Jiang SB (2010) Implementation and evaluation of various demons deformable image registration algorithms on a GPU. Phys Med Biol 55(1):207–219
Gu S, Chakraborty G, Champley K, Alessio AM, Claridge J, Rockne R, Muzi M, Krohn KA, Spence AM, Alvord EC, Anderson ARA, Kinahan PE, Swanson KR (2012) Applying a patient-specific bio-mathematical model of glioma growth to develop virtual [18F]-FMISO-PET images. Math Med Biol 29(1):31–48
Gunzburger MD (2003) Perspectives in flow control and optimization. SIAM, Philadelphia
Gurtin ME (1981) An introduction to continuum mechanics, Mathematics in Science and Engineering, vol 158. Academic Press, Cambridge
Ha L, Krüger J, Joshi S, Silva CT (2011) Multiscale unbiased diffeomorphic atlas construction on multi-GPUs. In: CPU computing gems Emerald edition, chap. 48. Elsevier Inc, New York City, pp 771–791
Ha LK, Krüger J, Fletcher PT, Joshi S, Silva CT (2009) Fast parallel unbiased diffeomorphic atlas construction on multi-graphics processing units. In: Proceedings of the eurographics conference on parallel grphics and visualization, pp 41–48 (2009)
Ha L, Krüger J, Joshi S, Silva TC (2010) Multi-scale unbiased diffeomorphic atlas construction on multi-GPUs. GPU Comput Gems Emerald Ed 1:771–791
Haber E, Ascher UM (2001) Preconditioned all-at-once methods for large, sparse parameter estimation problems. Inverse Probl 17(6):1847–1864
Haber E, Horesh R (2015) A multilevel method for the solution of time dependent optimal transport. Numer Math Theory Methods Appl 8(1):97–111
Haber E, Modersitzki J (2006) A multilevel method for image registration. SIAM J Sci Comput 27(5):1594–1607
Haber E, Ascher UM, Oldenburg D (2000) On optimization techniques for solving nonlinear inverse problems. Inverse Probl 16:1263–1280
Hager WW (2000) Runge-Kutta methods in optimal control and the transformed adjoint system. Numer Math 87:247–282
Hansen C (1992) Analysis of discrete ill-posed problems by means of the L-curve. SIAM Rev 34(4):561–580
Harpold HLP, Alvord EC, Swanson KR (2007) The evolution of mathematical modeling of glioma proliferation and invasion. J Neuropathol Exp Neurol 66(1):1–9
Hart GL, Zach C, Niethammer M (2009) An optimal control approach for deformable registration. In: Proceedings of the IEEE conference on computer vision and pattern recognition, pp 9–16
Hawkins-Daarud A, Prudhomme S, van der Zee KG, Oden JT (2013a) Bayesian calibration, validation, and uncertainty quantification of diffuse interface models of tumour growth. J Math Biol 67(6–7):1457–1485
Hawkins-Daarud A, Rockne RC, Anderson ARA, Swanson KR (2013b) Modeling tumor-associated edema in gliomas during anti-angiogenic therapy and its impact on imageable tumor. Front Oncol 3(66):1–12
Heinkenschloss M (2005) A time-domain decomposition iterative method for the solution of distributed linear quadratic optimal control problems. J Comput Appl Math 1(1):169–198
Hernandez M, Bossa MN, Olmos S (2009) Registration of anatomical images using paths of diffeomorphisms parameterized with stationary vector field flows. Int J Comput Vis 85(3):291–306
Herzog R, Kunisch K (2010) Algorithms for PDE-constrained optimization. GAMM Mitt 33(2):163–176
Herzog R, Pearson JW, Stoll M (2018) Fast iterative solvers for an optimal transport problem. arXiv: 1801.04172
Hinze M, Pinnau R, Ulbrich M, Ulbrich S (2009) Optimization with PDE constraints. Springer, Berlin
Hogea C, Davatzikos C, Biros G (2007) Modeling glioma growth and mass effect in 3D MR images of the brain. In: Proceedings of the medical image computing and computer-assisted intervention, pp 642–650
Hogea C, Davatzikos C, Biros G (2008a) Brain-tumor interaction biophysical models for medical image registration. SIAM J Imaging Sci 30(6):3050–3072
Hogea C, Davatzikos C, Biros G (2008b) An image-driven parameter estimation problem for a reaction-diffusion glioma growth model with mass effects. J Math Biol 56(6):793–825
Hormuth II DA, Weis JA, Barnes SL, Miga MI, Rericha EC, Quaranta V, Yankeelov TE (2015) Predicting in vivo glioma growth wit the reaction diffusion equation constrained by quantitative magnetic resonance imaging data. Phys Biol 12(4):046006
Horn BKP, Shunck BG (1981) Determining optical flow. Artif Intell 17(1–3):185–203
Hu Z, Metaxas D, Axel L (2003) In vivo strain and stress estimation of the heart left and right ventricles from MRI images. Med Image Anal 7(4):435–444
Jackson PR, Juliano J, Hawkins-Daarud A, Rockne RC, Swanson KR (2015) Patient-specific mathematical neuro-oncology: Using a simple proliferation and invasion tumor model to inform clinical practice. Bull Math Biol 77(5):846–856
Joshi A, Bangerth W, Sevick-Muraca EM (2004) Adaptive finite element based tomography for fluorescence optical imaging in tissue. Opt Express 12(22):5402–5417
Joshi S, Davis B, Jornier M, Gerig G (2005) Unbiased diffeomorphic atlas construction for computational anatomy. NeuroImage 23(1):S151–S160
Kaipio J, Somersalo E (2005) Statistical and computational inverse problems. Springer, Berlin
Kalmoun EM, Garrido L, Caselles V (2011) Line search multilevel optimization as computational methods for dense optical flow. SIAM J Imaging Sci 4(2):695–722
Klein S, Staring M, Murphy K, Viergever MA, Pluim JPW (2010) ELASTIX: a tollbox for intensity-based medical image registration. IEEE Trans Med Imaging 29(1):196–205
Knopoff DA, Fernández DR, Torres GA, Turner CV (2013) Adjoint method for a tumor growth PDE-constrained optimization problem. Comput Math Appl 66(6):1104–1119
Knopoff D, Fernández DR, Torres GA, Turner CV (2017) A mathematical method for parameter estimation in a tumor growth model. Comput Appl Math 36(1):733–748
Kø N, Tanderup K, Lindegaard JC, Grau C, Søorensen TS (2008) GPU accelerated viscous-fluid deformable registration for radiotherapy. Stud Health Technol Inform 132:327–332
Konukoglu E, Clatz O, Bondiau PY, Delingette H, Ayache N (2010) Extrapolating glioma invasion margin in brain magnetic resonance images: suggesting new irradiation margins. Med Image Anal 14(2):111–125
Konukoglu E, Clatz O, Menze BH, Stieltjes B, Weber MA, Mandonnet E, Delingette H, Ayache N (2010) Image guided personalization of reaction-diffusion type tumor growth models using modified anisotropic eikonal equations. IEEE Trans Med Imaging 29(1):77–95
Kutten KS, Charon N, Miller MI, Ratnanather JT, Deisseroth K, Ye L, Vogelstein JT (2017) A diffeomorphic approach to multimodal registration with mutual information: Applications to CLARITY mouse brain images. Proceedings of the medical image computing and computer-assisted intervention, vol LNCS 10433:275–282
Lê M, Delingette H, Kalpathy-Cramer J, Gerstner ER, Batchelor T, Unkelbach J, Ayache N (2015) Bayesian personalization of brain tumor growth model. In: Proceedings of the medical image computing and computer-assisted intervention, pp 424–432
Lê M, Delingette H, Kalpathy-Cramer J, Gerstner ER, Batchelor T, Unkelbach J, Ayache N (2017) Personalized radiotherapy planning based on a computational tumor growth model. IEEE Trans Med Imaging 36(3):815–825
Lee E, Gunzburger M (2010) An optimal control formulation of an image registration problem. J Math Imaging Vis 36(1):69–80
Lee E, Gunzburger M (2011) Anaysis of finite element discretization of an optimal control formulation of the image registration problem. SIAM J Numer Anal 9(4):1321–1349
Leugering G, Benner P, Engell S, Griewank A, Harbrecht H, Hinze M, Rannacher R, Ulbrich S (eds) (2014) Trends in PDE constrained optimization. Springer, Berlin
Lima E, Oden J, Almeida R (2014) A hybrid ten-species phase-field model of tumor growth. Math Models Methods Appl Sci 24(13):2569–2599
Lima EABF, Oden JT, Hormuth DA, Yankeelov TE, Almeida RC (2016) Selection, calibration, and validation of models of tumor growth. Math Models Methods Appl Sci 26(12):2341–2368
Lima E, Oden JT, Wohlmuth B, Shahmoradi A, Hormuth DA, Yankeelov TE (2017) Selection and validation of predictive models of radiation effects on tumor growth based on noninvasive imaging data. Comput Methods Appl Mech Eng 327:277–305
Lions JL (1971) Optimal control of systems governed by partial differential equations. Springer, Berlin
Liu Y, Sadowki SM, Weisbrod AB, Kebebew E, Summers RM, Yao J (2014) Patient specific tumor growth prediction using multimodal images. Med Image Anal 18(3):555–566
Luo Y, Liu P, Shi L, Luo Y, Yi L, Li A, Qin J, Heng PA, Wang D (2015) Accelerating neuroimage registration through parallel computation of similarity metric. PLoS ONE 10(9): e0136,718 (2015)
Mang A (2014) Methoden zur numerischen Simulation der Progression von Gliomen: Modellentwicklung. Springer Fachmedien Wiesbaden, Wiesbaden, Numerik und Parameteridentifikation. https://doi.org/10.1007/978-3-658-05246-1
Mang A, Biros G (2015) An inexact Newton-Krylov algorithm for constrained diffeomorphic image registration. SIAM J Imaging Sci 8(2):1030–1069. https://doi.org/10.1137/140984002
Mang A, Biros G (2016) Constrained \(H^1\)-regularization schemes for diffeomorphic image registration. SIAM J Imaging Sci 9(3):1154–1194. https://doi.org/10.1137/15M1010919
Mang A, Biros G (2017) A semi-Lagrangian two-level preconditioned Newton-Krylov solver for constrained diffeomorphic image registration. SIAM J Sci Comput 39(5):B860–B885. https://doi.org/10.1137/16M1070475
Mang A, Ruthotto L (2017) A Lagrangian Gauss–Newton–Krylov solver for mass- and intensity-preserving diffeomorphic image registration. SIAM J Sci Comput 39(5):B860–B885. https://doi.org/10.1137/17M1114132
Mang A, Schuetz TA, Becker S, Toma A, Buzug TM (2012a) Cyclic numerical time integration in variational non-rigid image registration based on quadratic regularisation. In: Proceedings of the vision, modeling and visualization workshop, pp 143–150. https://doi.org/10.2312/PE/VMV/VMV12/143-150
Mang A, Toma A, Schuetz TA, Becker S, Eckey T, Mohr C, Petersen D, Buzug TM (2012b) Biophysical modeling of brain tumor progression: from unconditionally stable explicit time integration to an inverse problem with parabolic PDE constraints for model calibration. Med Phys 39(7):4444–4459. https://doi.org/10.1118/1.4722749
Mang A, Gholami A, Biros G (2016) Distributed-memory large-deformation diffeomorphic 3D image registration. In: Proceedings of the ACM/IEEE conference on supercomputing, p 72. https://doi.org/10.1145/3126908.3126930
Mang A, Tharakan S, Gholami A, Nimthani N, Subramanian S, Levitt J, Azmat M, Scheufele K, Mehl M, Davatzikos C, Barth B, Biros G (2017) SIBIA-GlS: scalable biophysics-based image analysis for glioma segmentation. In: Proceedings of the BraTS 2017 workshop, pp 197–204
Martin J, Wilcox LC, Burstedde C, Ghattas O (2012) A stochastic Newton MCMC method for large-scale statistical inverse problems with application to seismic inversion. SIAM J Sci Comput 34(3):A1460–A1487
Menze BH, Jakab A, Bauer S, Kalpathy-Cramer J, Farahani K, Kirby J, Burren Y, Porz N, Slotboom J, Wiest R, Lanczi L, Gerstner E, Weber MA, Arbel T, Avants BB, Ayache N, Buendia P, Collins DL, Cordier N, Corso JJ, Criminisi A, Das T, Delingette H, Demiralp Ç, Durst CR, Dojat M, Doyle S, Festa J, Forbes F, Geremia E, Glocker B, Golland P, Guo X, Hamamci A, Iftekharuddin KM, Jena R, John NM, Konukoglu E, Lashkari D, Mariz JA, Meier R, Pereira S, Precup D, Price SJ, Raviv TR, Reza SMS, Ryan M, Sarikaya D, Schwartz L, Shin HC, Shotton J, Silva CA, Sousa N, Subbanna NK, Szekely G, Taylor TJ, Thomas OM, Tustison NJ, Unal G, Vasseur F, Wintermark M, Ye DH, Zhao L, Zhao B, Zikic D, Prastawa M, Reyes M, Leemput KV (2015) The multimodal brain tumor image segmentation benchmark (BRATS). IEEE Trans Med Imaging 34(10):1993–2024
Menze BH, Van Leemput K, Honkela A, Konukoglu E, Weber MA, Ayache N, Golland P (2011) A generative approach for image-based modeling of tumor growth. In: Information processing in medical imaging (IPMI 2011). Lecture notes in computer science, vol 6801. pp 735–747
Mi H, Petitjean C, Dubray B, Vera P, Ruan S (2014) Prediction of lung tumor evolution during radiotherapy in individual patients with PET. IEEE Trans Med Imaging 33(4):995–1003
Miller MI (2004) Computational anatomy: shape, growth and atrophy comparison via diffeomorphisms. NeuroImage 23(1):S19–S33
Miller MI, Younes L (2001) Group actions, homeomorphism, and matching: a general framework. Int J Comput Vis 41(1/2):61–81
Miller MI, Trouvé A, Younes L (2006) Geodesic shooting for computational anatomy. J Math Imaging Vis 24:209–228
Modat M, Ridgway GR, Taylor ZA, Lehmann M, Barnes J, Hawkes DJ, Fox NC, Ourselin S (2010) Fast free-form deformation using graphics processing units. Comput Methods Programs Biomed 98(3):278–284
Modersitzki J (2004) Numerical methods for image registration. Oxford University Press, New York
Modersitzki J (2009) FAIR: flexible algorithms for image registration. SIAM, Philadelphia
Mohamed A, Davatzikos C (2005) Finite element modeling of brain tumor mass-effect from 3D medical images. In: Proceedings of the medical image computing and computer-assisted intervention, pp 400–408
Mori S, Oishi K, Jiang H, Jiang L, Li X, Akhter K, Hua K, Faria AV, Mahmood A, Woods R, Toga AW, Pike GB, Neto PR, Evans A, Zhang J, Huang H, Miller MI, van Zijl P, Mazziotta J (2008) Stereotaxic white matter atlas based on diffusion tensor imaging in an ICBM template. NeuroImage 40(2):570–582
Mosayebi P, Cobzas D, Murtha A, Jagersand M (2012) Tumor invasion margin on the Riemannian space of brain fibers. Med Image Anal 16(2):361–373
Munson T, Sarich J, Wild S, Benson S, McInnes LC (2017) TAO 3.7 users manual. Argonne National Laboratory, Mathematics and Computer Science Division, Illinois
Murray JD (1989) Mathematical biology. Springer, New York
Muyan-Ozcelik P, Owens JD, Xia J, Samant SS (2008) Fast deformable registration on the GPU: A CUDA implementation of demons. In: IEEE international conference on computational sciences and its applications, pp 223–233
Nocedal J, Wright SJ (2006) Numerical optimization. Springer, New York
Oberai AA, Gokhale NH, Feijóo RG (2003) Solution of inverse problems in elasticity imaging using the adjoint method. Inverse Probl 19(2):297
Oden JT, Prudencio EE, Hawkins-Daarud A (2013) Selection and assessment of phenomenological models of tumor growth. Math Models Methods Appl Sci 23(7):1309–1338
Ophir J, Alam SK, Garra B, Kallel F, Konofagou E, Krouskop T, Varghese T (1999) Elastography: ultrasonic estimation and imaging of the elastic properties of tissues. J Eng Med 213(3):203–233
Ou Y, Sotiras A, Paragios N, Davatzikos C (2011) DRAMMS: deformable registration via attribute matching and mutual-saliency weighting. Med Image Anal 15(4):622–639
Papademetris X, Sinusas A, Dione D, Duncan J (2001) Estimation of 3D left ventricular deformation from echocardiography. Med Image Anal 5(1):17–28
Papademetris X, Sinusas AJ, Dione DP, Constable RT, Duncan JS (2002) Estimation of 3D left ventricular deformation from medical images using biomechanical models. IEEE Trans Med Imaging 21(7):786–800
Pearson JW, Stoll M (2013) Fast iterative solution of reaction-diffusion control problems arising from chemical processes. SIAM J Sci Comput 35(5):B987–B1009
Perperidis D, Mohiaddin R, Rueckert D (2005a) Construction of a 4D statistical atlas of the cardiac anatomy and its use in classification. In: Proceedings of the medical image computing and computer-assisted intervention (Lecture notes in computer science), vol 3750. Springer, Berlin, pp 402–410
Perperidis D, Mohiaddin RH, Rueckert D (2005b) Spatio-temporal free-form registration of cardiac MR image sequences. Med Image Anal 9(5):441–456
Petra N, Martin J, Stadler G, Ghattas O (2014) A computational framework for infinite-dimensional Bayesian inverse problems Part II: stochastic Newton MCMC with application to ice sheet flow inverse problems. SIAM J Sci Comput 36(4):A1525–A1555
Pock T, Urschler M, Zach C, Beichel R, Bischof H (2007) A duality based algorithm for TV-L\(^1\)-optical-flow image registration. Proceedings of the medical image computing and computer-assisted intervention, vol LNCS 4792:511–518
Powathil G, Kohandel M, Sivaloganathan S, Oza A, Milosevic M (2007) Mathematical modeling of brain tumors: effects of radiotherapy and chemotherapy. Phys Med Biol 52(11):3291
Quiroga AAI, Fernández D, Torres GA, Turner CV (2015) Adjoint method for a tumor invasion PDE-constrained optimization problem in 2D using adaptive finite element method. Appl Math Comput 270:358–368
Rahman MM, Feng Y, Yankeelov TE, Oden JT (2017) A fully coupled space-time multiscale modeling framework for predicting tumor growth. Comput Methods Appl Mech Eng 320:261–286
Rekik I, Allassonnière S, Clatz O, Geremia E, Stretton E, Delingette H, Ayache N (2013) Tumor growth parameters estimation and source localization from a unique time point: Application to low-grade gliomas. Comput Vis Image Underst 117(3):238–249
Ren K, Bal G, Hielscher AH (2006) Frequency domain optical tomography based on the equation of radiative transfer. SIAM J Sci Comput 28(4):1463–1489
Rockne R, Rockhill JK, Mrugala M, Spence AM, Kalet I, Hendrickson K, Lai A, Cloughesy T, Alvord EC, Swanson KR (2010) Predicting the efficacy of radiotherapy in individual glioblastoma patients in vivo: a mathematical modeling approach. Phys Med Biol 55(12):3271
Roose T, Chapman SJ, Maini PK (2007) Mathematical models of avascular tumor growth. SIAM Rev 49(2):179–208
Rühaak J, König L, Tramnitzke F, Köstler H, Modersitzki J (2017) A matrix-free approach to efficient affine-linear image registration on CPU and GPU. J Real Time Image Proc 13(1):205–225
Ruhnau P, Schnörr C (2007) Optical Stokes flow estimation: an imaging-based control approach. Exp Fluids 42:61–78
Saratoon T, Tarvainen, T, Cox BT, Arridge SR (2013) A gradient-based method for quantitative photoacoustic tomography using the radiative transfer equation. Inverse Probl 29(7): 075,006
Scheufele K, Mang A, Gholami A, Davatzikos C, Biros G, Mehl M (2018) Coupling brain-tumor biophysical models and diffeomorphic image registration. arXiv: 1710.06420
Schuetz TA, Becker S, Mang A, Toma A, Buzug TM (2013) Modelling of glioblastoma growth by linking a molecular interaction network with an agent based model. Math Comput Model Dyn Syst 19(5):417–433. https://doi.org/10.1080/13873954.2013.777748
Sermesant M, Delingette H, Ayache N (2006a) An electromechanical model of the heart for image analysis and simulation. IEEE Trans Med Imaging 25(5):612–625
Sermesant M, Moireau P, Camara O, Sainte-Marie J, Andriantsimiavona R, Cimrman R, Hill DL, Chapelle D, Razavi R (2006b) Cardiac function estimation from MRI using a heart model and data assimilation: advances and difficulties. Med Image Anal 10(4):642–656
Shackleford J, Kandasamy N, Sharp G (2013) High performance deformable image registration algorithms for manycore processors. Morgan Kaufmann, Waltham
Shah DJ, Judd RM, Kim RJ (2005) Technology insight: MRI of the myocardium. Nat Clin Pract Cardiovasc Med 2(11):597–605
Shams R, Sadeghi P, Kennedy R, Hartley R (2010a) Parallel computation of mutual information on the GPU with application to real-time registration of 3D medical images. Comput Methods Programs Biomed 99:133–146
Shams R, Sadeghi P, Kennedy RA, Hartley RI (2010b) A survey of medical image registration on multicore and the GPU. Signal Process Mag IEEE 27(2):50–60
Shen DG, Sundar H, Xue Z, Fan Y, Litt H (2005) Consistent estimation of cardiac motions by 4D image registration. In: Proceedings of the Medical image computing and computer-assisted intervention (Lecture notes in computer science), vol 3750. Springer, Berlin, pp 902–910
Shenk O, Manguoglu M, Sameh A, Christen M, Sathe M (2009) Parallel scalable PDE-constrained optimization: antenna identification in hyperthermia cancer treatment planning. Comput Sci Res Dev 23(3–4):177–183
Simoncini V (2012) Reduced order solution of structured linear systems arising in certain PDE-constrained optimization problems. Comput Optim Appl 53(2):591–617
Sommer S (2008) Accelerating multi-scale flows for LDDKBM diffeomorphic registration. In: Proceedings of the IEEE international conference on computer visions workshops, pp 499–505
Sotiras A, Davatzikos C, Paragios N (2013) Deformable medical image registration: a survey. IEEE Trans Med Imaging 32(7):1153–1190
Sullivan TJ (2015) Introduction to uncertainty quantification. Springer, Berlin
Sundar H, Davatzikos C, Biros G (2009) Biomechanically constrained 4D estimation of mycardial motion. In: Proceedings of the medical image computing and computer-assisted intervention, vol LNCS 5762, pp 257–265
Swanson KR, Alvord EC, Murray JD (2000) A quantitative model for differential motility of gliomas in grey and white matter. Cell Prolif 33(5):317–330
Swanson KR, Alvord EC, Murray JD (2002) Virtual brain tumours (gliomas) enhance the reality of medical imaging and highlight inadequacies of current therapy. Br J Cancer 86(1):14–18
Swanson KR, Rostomily RC, Alvord EC (2008) A mathematical modelling tool for predicting survival of individual patients following resection of glioblastoma: a proof of principle. Br J Cancer 98(1):113–119
Tarantola A (2005) Inverse problem theory and methods for model parameter estimation. SIAM, Philadelphia
Toma A, Mang A, Schuetz TA, Becker S, Buzug TM (2012) A novel method for simulating the extracellular matrix in models of tumour growth. Comput Math Methods Med 2012:960,256-1–960,256-11. https://doi.org/10.1155/2012/109019
Tomer R, Ye L, Hsueh B, Deisseroth K (2014) Advanced CLARITY for rapid and high-resolution imaging of intact tissues. Nat Protoc 9(7):1682–1697
Trouvé A (1998) Diffeomorphism groups and pattern matching in image analysis. Int J Comput Vis 28(3):213–221
Tuyisenge V, Sarry L, Corpetti T, Innorta-Coupez E, Ouchchane L, Cassagnes L (2016) Estimation of myocardial strain and contraction phase from cine MRI using variational data assimilation. IEEE Trans Med Imag 35(2):442–455
ur Rehman T, Haber E, Pryor G, Melonakos J, Tannenbaum A (2009) 3D nonrigid registration via optimal mass transport on the GPU. Med Image Anal 13(6):931–940
Valero-Lara P (2014) Multi-GPU acceleration of DARTEL (early detection of Alzheimer). In: Proceedings of the IEEE international conference on cluster computing, pp 346–354
Vercauteren T, Pennec X, Perchant A, Ayache N (2008) Symmetric log-domain diffeomorphic registration: a demons-based approach. Proceedings of the medical image computing and computer-assisted intervention, vol LNCS 5241:754–761
Vercauteren T, Pennec X, Perchant A, Ayache N (2009) Diffeomorphic demons: efficient non-parametric image registration. NeuroImage 45(1):S61–S72
Vialard FX, Risser L, Rueckert D, Cotter CJ (2012) Diffeomorphic 3D image registration via geodesic shooting using an efficient adjoint calculation. Int J Comput Vis 97:229–241
Wang Z, Deisboeck TS (2008) Computational modeling of brain tumors: discrete, continuum or hybrid. Sci Model Simul SMNS 15(1–3):381
Weis JA, Miga MI, Yankeelov TE (2017) Three-dimensional image-based mechanical modeling for predicting the response of breast cancer to neoadjuvant therapy. Comput Methods Appl Mech Eng 314:494–512
Wilcox LC, Stadler G, Bui-Thanh T, Ghattas O (2015) Discretely exact derivatives for hyperbolic PDE-constrained optimization problems discretized by the discontinuous Galerkin method. J Sci Comput 63(1):138–162
Wlazło J, Fessler R, Pinnau R, Siedow N, Tse O (2016) Elastic image registration with exact mass preservation. arXiv: 1609.04043
Wong KCL, Summers RM, Kebebew E, Yao J (2015) Pancreatic tumor growth prediction with multiplicative growth and image-derived motion. Proceedings of the information processing in medical imaging, vol LNCS 9123:501–513
Wong KCL, Summers RM, Kebebew E, Yoa J (2017) Pancreatic tumor growth prediction with elastic-growth decomposition, image-derived motion, and FDM-FEM coupling. IEEE Trans Med Imaging 36(1):111–123
Younes L (2007) Jacobi fields in groups of diffeomorphisms and applications. Q Appl Math 650(1):113–134
Younes L (2010) Shapes and diffeomorphisms. Springer, Berlin
Zacharaki EI, Hogea CS, Biros G, Davatzikos C (2008a) A comparative study of biomechanical simulators in deformable registration of brain tumor images. IEEE Trans Biomed Eng 55(3):1233–1236
Zacharaki EI, Hogea CS, Shen D, Biros G, Davatzikos C (2008b) Parallel optimization of tumor model parameters for fast registration of brain tumor images. In: Proceedings of the SPIE medical imaging, pp 69,140K1–69,140K10
Zacharaki EI, Hogea CS, Shen D, Biros G, Davatzikos C (2009) Non-diffeomorphic registration of brain tumor images by simulating tissue loss and tumor growth. NeuroImage 46(3):762–774
Zhang M, Fletcher PT (2015) Bayesian principal geodesic analysis for estimating intrinsic diffeomorphic image variability. Med Image Anal 25(1):37–44
Acknowledgements
The authors thank the anonymous reviewers for their careful reading and their insightful comments.
Funding
This material is based upon work supported by AFOSR grants FA9550-17-1-0190; by NSF Grant CCF-1337393; by the U.S. Department of Energy, Office of Science, Office of Advanced Scientific Computing Research, Applied Mathematics program under Award Numbers DE-SC0010518 and DE-SC0009286; by NIH Grant 10042242; by DARPA Grant W911NF-115-2-0121; and by the Technische Universität München—Institute for Advanced Study, funded by the German Excellence Initiative (and the European Union Seventh Framework Programme under grant agreement 291763). Any opinions, findings, and conclusions or recommendations expressed herein are those of the authors and do not necessarily reflect the views of the AFOSR, DOE, NIH, DARPA, and NSF. Computing time on the Texas Advanced Computing Center’s (TACC) systems was provided by an allocation from TACC and the NSF. Computing time on the High-Performance Computing Center’s (HLRS) Hazel Hen system (Stuttgart, Germany) was provided by an allocation of the federal project application ACID-44104. This work was completed in part with resources provided by the University of Houston Center for Advanced Computing and Data Science (CACDS)
Author information
Authors and Affiliations
Corresponding author
Appendices
Appendix 1: Computing \(\det \nabla y\)
We do not differentiate the deformation map y, but transport \(\psi \mathrel {\mathop :}=\det \nabla \phi\) where \(y^{-1}(x) = \phi (x,t=1)\). That is, we solve
with periodic boundary conditions on \(\partial {\varOmega }\).
Remark 6
If we start from an identity map \({\text {id}}\) (zero velocity field) the determinant of the deformation gradient will have a value of one. Hence, we require that the determinant of the deformation gradient remains strictly positive for all \(x \in {\varOmega }\) for the computed deformation map to be locally diffeomorphic. Strictly speaking, we also require that the map y is smooth, and has a smooth inverse for y to be a diffeomorphism. If we use (slightly more than) \(H^2\)-regularity for v we meet these requirements from a theoretical perspective (assuming that the images are smooth) (Beg et al. 2005; Vialard et al. 2012). However, we note that ensuring that the discrete deformation map y (which does not appear in our formulation) is a diffeomorphism, requires sophisticated discretization schemes (see, e.g., Burger et al. 2013). In our formulation, we transport the data. Since the deformation map y does not appear explicitly, we decided to compute the determinant of the deformation gradient by computing the solution of the transport problem in Appendix 1. We found that this is more stable than computing \(\det \nabla y\) from y using our numerical scheme.
Appendix 2: Gauss–Newton Krylov algorithm
Here, we showcase the individual steps necessary for solving (1) in Algorithm 3 (Newton iterations; outer iterations) and Algorithm 4 (solution of the system in (12), inner iterations). We refer to Mang and Biros (2015, 2017), Gholami et al. (2016) for details on the particular implementation of our nonlinear solver. We rely on PETSc’s TAO package (Balay et al. 2016; Munson et al. 2017) for our distributed-memory solver (Mang et al. 2016; Gholami et al. 2017) (see Sect. 5.5 for details). The steps are the same as outlined in Algorithms 3 and 4, respectively.


Appendix 3: Newton step
Here, we describe the Newton step for the two problems described in Sects. 2.2 and 3.2. The Newton step is obtained by computing the second variation of (6). We will see that the structure of the PDE operators that appear in the reduced space Hessian is very similar to the first-order optimality systems in Sect. 5.1.
1.1 Diffeomorphic registration
The nonlinearity, ill-posedness, and the dimensionality of the search space makes the solution of this variational problem computational challenging. Most available implementations use first-order iterative schemes to compute a minimizer to the variational optimization problem (see, e.g., Avants et al. 2011; Beg et al. 2005; Chen and Lorenz 2011; Ha et al. 2010; Hart et al. 2009; Vialard et al. 2012). To increase the rate of convergence, they typically do not use \(g_v\) in their iterative scheme, but the gradient in the Sobolev space induced by the regularization operator \({\mathcal {A}}\), i.e.,
First-order methods are usually inefficient; they require a lot of iterations to converge to a specific tolerance; we have analyzed this in Mang and Biros (2015). An alternative is to apply variants of Newton’s method to the KKT system. To derive the operators for applying Newton’s method we have to compute the second variations of (6). We arrive at the incremental state and adjoint equation
and the expression for the reduced space Hessian matvec
with periodic boundary conditions on \(\partial {\varOmega }\). The incremental control variable \(\tilde{v}\) corresponds to the search direction of our scheme. The transport equations for the incremental state and adjoint fields \(\tilde{m}\) and \(\tilde{\lambda }\) are hidden in the integro-differential operator in (14e); we have to solve (14a) and (14c) every time we apply \({\mathcal {H}}\) to a new vector \(\tilde{v}\). We use a Gauss–Newton approximation to the true Hessian to ensure that the operator is positive definite far away from the optimum. This corresponds to neglecting all terms that involve \(\lambda\) in (14c) and (14e).
1.2 Data assimilation
The expressions for evaluating the Hessian operator are derived by computing the second variations for (9). The Hessian matvec is given by \({\mathcal {H}}\tilde{p} \mathrel {\mathop :}=\beta \tilde{p} - {\varPhi }^\mathsf {T}\tilde{\lambda }_0\), \({\mathcal {H}} : {\mathbf {R}}^{n_p} \rightarrow {\mathbf {R}}^{n_p}\). To be able to evaluate this expression we have to solve the incremental state and adjoint equations given by
with Neumann boundary conditions on \(\partial {\varOmega }_B\). For the Gauss–Newton approximation to the true Hessian the terms that involve \(\lambda\) in (15c) need to be dropped.
Rights and permissions
About this article
Cite this article
Mang, A., Gholami, A., Davatzikos, C. et al. PDE-constrained optimization in medical image analysis. Optim Eng 19, 765–812 (2018). https://doi.org/10.1007/s11081-018-9390-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11081-018-9390-9