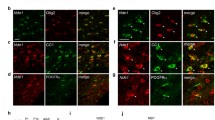
Abstract
Dystonia musculorum (dt) mice show sensory neurodegeneration and movement disorder, such as dystonia and cerebellar ataxia. The causative gene Dystonin (Dst) encodes a cytoskeleton linker protein. Although sensory neurodegeneration has been well studied, glial cell responses in the central nervous system (CNS) are poorly understood. Here, we investigated cell proliferation in the CNS of DstGt homozygous mice using newly generated in situ hybridization (ISH) probes—Ki-67 and proliferating cell nuclear antigen (PCNA) probes—both of which effectively detect proliferating cells. We found that Ki-67-positive cells were significantly decreased in the corpus callosum and thalamus of dt brain at postnatal day 21 (P21). There is a similar but not significant tendency at postnatal day 14 (P14) in the dt brain. We also confirmed the reduced proliferation by PCNA ISH and Ki-67 immunohistochemistry. Double staining with cell-type-specific markers revealed that proliferating cells are oligodendrocyte progenitor cells (OPCs) in both wild-type and dt brain. We also observed a reduced number of Olig2-positive cells in the corpus callosum of DstGt homozygous mice at P21, indicating that reduced proliferation resulted in a reduced number of OPCs. Our data indicate that OPCs proliferation is reduced in the dt mouse brain at the postnatal stage and that it subsequently results in the reduced number of OPCs.





Similar content being viewed by others
References
Woodruff RH, Tekki-Kessaris N, Stiles CD, Rowitch DH, Richardson WD (2001) Int J Dev Neurosci 19:379–385
Thomas JL, Spassky N, Perez Villegas EM, Olivier C, Cobos I, Goujet-Zalc C, Martínez S, Zalc B (2000) Spatiotemporal development of oligodendrocytes in the embryonic brain. J Neurosci Res 59:471–476
Takebayashi H, Ikenaka K (2015) Oligodendrocyte generation during mouse development. Glia 63:1350–1356. doi:https://doi.org/10.1002/glia.22863
Ono K, Takebayashi H, Ikeda K, Furusho M, Nishizawa T, Watanabe K, Ikenaka K (2008) Regional- and temporal-dependent changes in the differentiation of Olig2 progenitors in the forebrain, and the impact on astrocyte development in the dorsal pallium. Dev Biol 320:456–468. doi:https://doi.org/10.1016/j.ydbio.2008.06.001
Cayre M, Canoll P, Goldman JE (2009) Cell migration in the normal and pathological postnatal mammalian brain. Prog Neurobiol 88:41–63. doi:https://doi.org/10.1016/j.pneurobio.2009.02.001
Psachoulia K, Jamen F, Young KM, Richardson WD (2009) Cell cycle dynamics of NG2 cells in the postnatal and ageing brain. Neuron Glia Biol 5:57–67. doi:https://doi.org/10.1017/S1740925X09990354
Pringle NP, Richardson WD. (1993) A singularity of PDGF alpha-receptor expression in the dorsoventral axis of the neural tube may define the origin of the oligodendrocyte lineage. Development 117:525–533
Stallcup WB, Beasley L (1987) Bipotential glial precursor cells of the optic nerve express the NG2 proteoglycan. J Neurosci 7:2737–2744
Diers-Fenger M, Kirchhoff F, Kettenmann H, Levine JM, Trotter J (2001) AN2/NG2 protein-expressing glial progenitor cells in the murine CNS: isolation, differentiation, and association with radial glia. Glia 34:213–228
Hill RA, Patel KD, Goncalves CM, Grutzendler J, Nishiyama A (2014) Modulation of oligodendrocyte generation during a critical temporal window after NG2 cell division. Nat Neurosci 17:1518–1527. doi:https://doi.org/10.1038/nn.3815
Lu QR, Yuk D, Alberta JA, Zhu Z, Pawlitzky I, Chan J, McMahon AP, Stiles CD, Rowitch DH (2000) Sonic hedgehog–regulated oligodendrocyte lineage genes encoding bHLH proteins in the mammalian central nervous system. Neuron 25:317–329
Takebayashi H, Yoshida S, Sugimori M, Kosako H, Kominami R, Nakafuku M, Nabeshima Y (2000) Dynamic expression of basic helix-loop-helix Olig family members: implication of Olig2 in neuron and oligodendrocyte differentiation and identification of a new member, Olig3. Mech Dev 99:143–148
Zhou Q, Wang S, Anderson DJ (2000) Identification of a novel family of oligodendrocyte lineage-specific basic helix-loop-helix transcription factors. Neuron 25:331–343
Gerdes J, Schwab U, Lemke H, Stein H (1983) Production of a mouse monoclonal antibody reactive with a human nuclear antigen associated with cell proliferation. Int J Cancer 31:13–20
Scholzen T, Gerdes J (2000) The Ki-67 protein: from the known and the unknown. J Cell Physiol 182:311–322
Muskhelishvili L, Latendresse JR, Kodell RL, Henderson EB (2003) Evaluation of cell proliferation in rat tissues with BrdU, PCNA, Ki-67(MIB-5) immunohistochemistry and in situ hybridization for histone mRNA. J Histochem Cytochem 51:1681–1688. doi:https://doi.org/10.1177/002215540305101212
Hall PA, Levison DA, Woods AL, Yu CC, Kellock DB, Watkins JA, Barnes DM, Gillett CE, Camplejohn R, Dover R, Waseem NH, Lane DP (1990) Proliferating cell nuclear antigen (PCNA) immunolocalization in paraffin sections: an index of cell proliferation with evidence of deregulated expression in some neoplasms. J Pathol 162:285–294
Moldovan GL, Pfander B, Jentsch S (2007) PCNA, the maestro of the replication fork. Cell 129:665–679. doi:https://doi.org/10.1016/j.cell.2007.05.003
Köhler T, Pröls F, Brand-Saberi B (2005) PCNA in situ hybridization: a novel and reliable tool for detection of dynamic changes in proliferative activity. Histochem Cell Biol 123:315–327. doi:https://doi.org/10.1007/s00418-004-0730-9
Duchen LW, Falconer DS, Strich SJ (1963) Dystonia musculorum: a hereditary neuropathy of mice affecting mainly sensory pathways. J Physiol 165:7–9
Janota I (1972) Ultrastructural studies of a hereditary sensory neuropathy in mice (dystonia musculorum). Brain 95:529–536
Thornburg LP, Hanker JS (1975) Lysosomal hydrolases in the trigeminal ganglion of mice afflicted with an hereditary sensory neuropathy (dystonia musculorum). Acta Neuropathol 32:91–101
Douglas DS, Popko B (2009) Mouse forward genetics in the study of the peripheral nervous system and human peripheral neuropathy. Neurochem Res 34:124–137. doi:https://doi.org/10.1007/s11064-008-9719-4
Guo L, Degenstein L, Dowling J, Yu QC, Wollmann R, Perman B, Fuchs E (1995) Gene targeting of BPAG1: abnormalities in mechanical strength and cell migration in stratified epithelia and neurologic degeneration. Cell 81:233–243
Brown A, Bernier G, Mathieu M, Rossant J, Kothary R (1995) The mouse dystonia musculorum gene is a neural isoform of bullous pemphigoid antigen 1. Nat Genet 10:301–306. doi:https://doi.org/10.1038/ng0795-301
Yang Y, Dowling J, Yu QC, Kouklis P, Cleveland DW, Fuchs E (1996) An essential cytoskeletal linker protein connecting actin microfilaments to intermediate filaments. Cell 86:655–665
Sonnenberg A, Liem RK (2007) Plakins in development and disease. Exp Cell Res 313:2189–2203. doi:https://doi.org/10.1016/j.yexcr.2007.03.039
Young KG, Kothary R (2007) Dystonin/Bpag1-A link to what? Cell Motil Cytoskeleton 64:897–905. doi:https://doi.org/10.1002/cm.20235
Horie M, Watanabe K, Bepari AK, Nashimoto J, Araki K, Sano H, Chiken S, Nambu A, Ono K, Ikenaka K, Kakita A, Yamamura K, Takebayashi H (2014) Disruption of actin-binding domain-containing Dystonin protein causes dystonia musculorum in mice. Eur J Neurosci 40:3458–3471. doi:https://doi.org/10.1111/ejn.12711
Edvardson S, Cinnamon Y, Jalas C, Shaag A, Maayan C, Axelrod FB, Elpeleg O (2012) Hereditary sensory autonomic neuropathy caused by a mutation in dystonin. Ann Neurol 71:569–572. doi:https://doi.org/10.1002/ana.23524
Horie M, Mekada K, Sano H, Kikkawa Y, Chiken S, Someya T, Saito K, Hossain MI, Nameta M, Abe K, Sakimura K, Ono K, Nambu A, Yoshiki A, Takebayashi H (2016) Characterization of novel dystonia musculorum mutant mice: implications for central nervous system abnormality. Neurobiol Dis 96:271–283. doi:https://doi.org/10.1016/j.nbd.2016.09.016
Seehusen F, Kiel K, Jottini S, Wohlsein P, Habierski A, Seibel K, Vogel Y, Urlaub H, Kollmar M, Baumgärtner W, Teichmann U (2016) Axonopathy in the central nervous system is the hallmark of mice with a novel intragenic null mutation of dystonin. Genetics 204:191–203. doi:https://doi.org/10.1534/genetics.116.186932
Kornfeld SF, Lynch-Godrei A, Bonin SR, Gibeault S, De Repentigny Y, Kothary R (2016) Cytoskeletal linker protein dystonin is not critical to terminal oligodendrocyte differentiation or CNS myelination. PLoS ONE 11:e0149201. doi:https://doi.org/10.1371/journal.pone.0149201
Ding L, Takebayashi H, Watanabe K, Ohtsuki T, Tanaka KF, Nabeshima Y, Chisaka O, Ikenaka K, Ono K (2005) Short-term lineage analysis of dorsally derived Olig3 cells in the developing spinal cord. Dev Dyn 234:622–632
Takebayashi H, Nabeshima Y, Yoshida S, Chisaka O, Ikenaka K, Nabeshima Y (2002) The basic helix-loop-helix factor olig2 is essential for the development of motoneuron and oligodendrocyte lineages. Curr Biol 12:1157–1163
Yokoo H, Nobusawa S, Takebayashi H, Ikenaka K, Isoda K, Kamiya M, Sasaki A, Hirato J, Nakazato Y (2004) Anti-human Olig2 antibody as a useful immunohistochemical marker of normal oligodendrocytes and gliomas. Am J Pathol 164:1717–1725. doi:https://doi.org/10.1016/S0002-9440(10)63730-3
Yamamura T, Konola JT, Wekerle H, Lees MB (1991) Monoclonal antibodies against myelin proteolipid protein: identification and characterization of two major determinants. J Neurochem 57:1671–1680. doi:https://doi.org/10.1111/j.1471-4159.1991.tb06367.x
Saulnier R, De Repentigny Y, Yong VW, Kothary R (2002) Alterations in myelination in the central nervous system of dystonia musculorum mice. J Neurosci Res 69:233–242. doi:https://doi.org/10.1002/jnr.10289
Rinholm JE, Hamilton NB, Kessaris N, Richardson WD, Bergersen LH, Attwell D (2011) Regulation of oligodendrocyte development and myelination by glucose and lactate. J Neurosci 31:538–548. doi:https://doi.org/10.1523/JNEUROSCI.3516-10.2011
Cahoy JD, Emery B, Kaushal A, Foo LC, Zamanian JL, Christopherson KS, Xing Y, Lubischer JL, Krieg PA, Krupenko SA, Thompson WJ, Barres BA (2008) A transcriptome database for astrocytes, neurons, and oligodendrocytes: a new resource for understanding brain development and function. J Neurosci 28:264–278. doi:https://doi.org/10.1523/JNEUROSCI.4178-07.2008
Bergles DE, Richardson WD (2015) Oligodendrocyte Development and Plasticity. Cold Spring Harb Perspect Biol 8:a020453. doi:https://doi.org/10.1101/cshperspect.a020453
Hof PR, Haroutunian V, Copland C, Davis KL, Buxbaum JD (2002) Molecular and cellular evidence for an oligodendrocyte abnormality in schizophrenia. Neurochem Res 27:1193–1200
Acknowledgements
This study was supported by JSPS KAKENHI Grant Numbers (JP15H04667, JP16K15168, JP24700351), Grant-in-Aid for Scientific Research on Innovative Areas, “Glial assembly” (JP25117007), the Cooperative Study Program of National Institute for Physiological Sciences, and a Grant from the Niigata University Kyowakai Society (IH). HT would like to thank Prof. Kaz Ikenaka for his continuing mentorship. We thank Prof. Marjorie B Lees and Prof. Kaz Ikenaka for AA3 antibody, Dr. Yukiko Mori, Mr. Yuya Imada, and Ms. Satoko Yamagiwa for technical assistance, and all members of Takebayashi lab, especially Dr. Nozomu Yoshioka for suggestions and advice.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
All authors declared no conflicts of interests with respect to the research, authorship, and/or publication of this article.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary Fig. 1
Reduced number of Ki-67-positive cells in the brain of DstGt homozygous mice at P21. a Ki-67 IHC of wild type (WT) and DstGt homozygotes (DstGt homo) was performed on the coronal sections. Representative images of corpus callosum and thalamus are shown. Red arrowheads indicate Ki-67-positive cells. b Double staining of Ki-67 ISH and Ki-67 IHC was performed on the coronal sections of wild type (WT) and DstGt homozygotes (DstGt homo). Representative images of corpus callosum and thalamus are shown. Black rectangles are shown at high magnification in insets. Red arrowheads indicate double positive cells and brown arrowheads indicate Ki-67 IHC single positive cells. Scale bars 100 µm, 20 µm (insets) (PPTX 357 KB)
Supplementary Fig. 2
Ki-67 IHC and PCNA ISH in the brain of wild-type and DstGt homozygous mice at P14. a, b Ki-67 IHC (a) and PCNA ISH (b) of wild-type mice and DstGt homozygotes were performed on coronal sections. After PCNA ISH, counterstaining was performed using nuclear fast red. Red arrowheads indicate positive signals. Scale bars 100 µm (a, b) (PPTX 358 KB)
Rights and permissions
About this article
Cite this article
Hossain, M.I., Horie, M. & Takebayashi, H. Reduced Proliferation of Oligodendrocyte Progenitor Cells in the Postnatal Brain of Dystonia Musculorum Mice. Neurochem Res 43, 101–109 (2018). https://doi.org/10.1007/s11064-017-2342-5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11064-017-2342-5