Abstract
Purpose
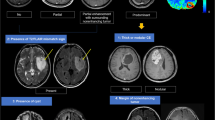
There is a growing body of literature documenting glioma heterogeneity in terms of radiographic, histologic, molecular, and genetic characteristics. Incomplete spatial specification of intraoperative tumor samples may contribute to variability in the results of pathological and biological investigations. We have developed a system, termed geo-tagging, for routine intraoperative linkage of each tumor sample to its location via neuronavigation.
Methods
This is a single-institution, IRB approved, prospective database of undergoing clinically indicated surgery. We evaluated relevant factors affecting data collection by this registry, including tumor and surgical factors (e.g. tumor volume, location, grade and surgeon).
Results
Over a 2-year period, 487 patients underwent craniotomy for an intra-axial tumor. Of those, 214 underwent surgery for a newly diagnosed or recurrent glioma. There was significant variation in the average number of samples collected per registered case, with a range of samples from 2.53 to 4.75 per tumor type. Histology and grade impacted on sampling with a range of 2.0 samples per tumor in Grade four, IDH-WT gliomas to 4.5 samples in grade four, IDH-mutant gliomas. The range of cases with sampling per surgeon was 6 to 99 with a mean of 47.6 cases and there was a statistically significant differences between surgeons. Tumor grade did not have a statistically significant impact on number of samples per case. No significant correlation was found between the number of samples collected and enhancing tumor volume, EOR, or volume of tumor resected.
Conclusion
We are using the results of this analysis to develop a prospective sample collection protocol.


Similar content being viewed by others
Data Availability
Deidentified data may be made available upon request.
References
Louis DN, Perry A, Wesseling P, Brat DJ, Cree IA, Figarella-Branger D, Hawkins C, Ng HK, Pfister SM, Reifenberger G, Soffietti R, von Deimling A, Ellison DW (2021) The 2021 WHO classification of tumors of the central nervous system: a summary. Neuro Oncol 23:1231–1251. https://doi.org/10.1093/neuonc/noab106
DeCordova S, Shastri A, Tsolaki AG, Yasmin H, Klein L, Singh SK, Kishore U (2020) Molecular heterogeneity and immunosuppressive microenvironment in glioblastoma. Front Immunol 11:1402. https://doi.org/10.3389/fimmu.2020.01402
Witthayanuwat S, Pesee M, Supaadirek C, Supakalin N, Thamronganantasakul K, Krusun S (2018) Survival analysis of glioblastoma multiforme. Asian Pac J Cancer Prev 19:2613–2617. https://doi.org/10.22034/APJCP.2018.19.9.2613
Becker AP, Sells BE, Haque SJ, Chakravarti A (2021) Tumor heterogeneity in glioblastomas: from light microscopy to molecular pathology. Cancers (Basel). https://doi.org/10.3390/cancers13040761
Lauko A, Lo A, Ahluwalia MS, Lathia JD (2022) Cancer cell heterogeneity & plasticity in glioblastoma and brain tumors. Semin Cancer Biol 82:162–175. https://doi.org/10.1016/j.semcancer.2021.02.014
Phillips HS, Kharbanda S, Chen R, Forrest WF, Soriano RH, Wu TD, Misra A, Nigro JM, Colman H, Soroceanu L, Williams PM, Modrusan Z, Feuerstein BG, Aldape K (2006) Molecular subclasses of high-grade glioma predict prognosis, delineate a pattern of disease progression, and resemble stages in neurogenesis. Cancer Cell 9:157–173. https://doi.org/10.1016/j.ccr.2006.02.019
Verhaak RG, Hoadley KA, Purdom E, Wang V, Qi Y, Wilkerson MD, Miller CR, Ding L, Golub T, Mesirov JP, Alexe G, Lawrence M, O’Kelly M, Tamayo P, Weir BA, Gabriel S, Winckler W, Gupta S, Jakkula L, Feiler HS, Hodgson JG, James CD, Sarkaria JN, Brennan C, Kahn A, Spellman PT, Wilson RK, Speed TP, Gray JW, Meyerson M, Getz G, Perou CM, Hayes DN, Cancer Genome Atlas Research N (2010) Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell 17:98–110. https://doi.org/10.1016/j.ccr.2009.12.020
Patel AP, Tirosh I, Trombetta JJ, Shalek AK, Gillespie SM, Wakimoto H, Cahill DP, Nahed BV, Curry WT, Martuza RL, Louis DN, Rozenblatt-Rosen O, Suva ML, Regev A, Bernstein BE (2014) Single-cell RNA-seq highlights intratumoral heterogeneity in primary glioblastoma. Science 344:1396–1401. https://doi.org/10.1126/science.1254257
Klughammer J, Kiesel B, Roetzer T, Fortelny N, Nemc A, Nenning KH, Furtner J, Sheffield NC, Datlinger P, Peter N, Nowosielski M, Augustin M, Mischkulnig M, Strobel T, Alpar D, Erguner B, Senekowitsch M, Moser P, Freyschlag CF, Kerschbaumer J, Thome C, Grams AE, Stockhammer G, Kitzwoegerer M, Oberndorfer S, Marhold F, Weis S, Trenkler J, Buchroithner J, Pichler J, Haybaeck J, Krassnig S, Mahdy Ali K, von Campe G, Payer F, Sherif C, Preiser J, Hauser T, Winkler PA, Kleindienst W, Wurtz F, Brandner-Kokalj T, Stultschnig M, Schweiger S, Dieckmann K, Preusser M, Langs G, Baumann B, Knosp E, Widhalm G, Marosi C, Hainfellner JA, Woehrer A, Bock C (2018) The DNA methylation landscape of glioblastoma disease progression shows extensive heterogeneity in time and space. Nat Med 24:1611–1624. https://doi.org/10.1038/s41591-018-0156-x
Comba A, Faisal SM, Varela ML, Hollon T, Al-Holou WN, Umemura Y, Nunez FJ, Motsch S, Castro MG, Lowenstein PR (2021) Uncovering spatiotemporal heterogeneity of high-grade gliomas: from disease biology to therapeutic implications. Front Oncol 11:703764. https://doi.org/10.3389/fonc.2021.703764
Weller M, Cloughesy T, Perry JR, Wick W (2013) Standards of care for treatment of recurrent glioblastoma—are we there yet? Neuro Oncol 15:4–27. https://doi.org/10.1093/neuonc/nos273
Jaoude DA, Moore JA, Moore MB, Twumasi-Ankrah P, Ablah E, Moore DF Jr (2019) Glioblastoma and increased survival with longer chemotherapy duration. Kans J Med 12:65–69
Olar A, Aldape KD (2014) Using the molecular classification of glioblastoma to inform personalized treatment. J Pathol 232:165–177. https://doi.org/10.1002/path.4282
Barajas RF Jr, Phillips JJ, Parvataneni R, Molinaro A, Essock-Burns E, Bourne G, Parsa AT, Aghi MK, McDermott MW, Berger MS, Cha S, Chang SM, Nelson SJ (2012) Regional variation in histopathologic features of tumor specimens from treatment-naive glioblastoma correlates with anatomic and physiologic MR imaging. Neuro Oncol 14:942–954. https://doi.org/10.1093/neuonc/nos128
Funding
The authors declare that no funds, grants or other support were received during the preparation of this manuscript.
Author information
Authors and Affiliations
Contributions
All authors contributed to the study conception and design. Material preparation, data collection and analysis were performed by JL, ZT and MAV. Initial drafting of the manuscript was performed by JL and MAV. All authors commented on previous versions of the manuscript, and have read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors have no financial interests that are relevant to the subject matter of this work. MAV has financial interests that are unrelated to this work, and include honoraria from Olympus, Midatech, and Chimerix.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Lynes, J., Khan, I., Aguilera, C. et al. Development of a “Geo-Tagged” tumor sample registry: intra-operative linkage of sample location to imaging. J Neurooncol 165, 449–458 (2023). https://doi.org/10.1007/s11060-023-04493-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11060-023-04493-2